Figure 2.
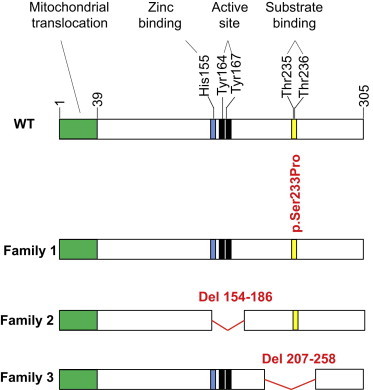
Effect of Genetic Variants Identified in CA-VA
Shown is a schematic diagram of the 305 amino acid wild-type (WT) CA-VA. Residues 1–39 encode a mitochondrial translocation signal (green). Homology predictions indicate that histidine 155 binds a zinc ion (blue), tyrosines 164 and 167 are active site residues (black), and threonines 235 and 236 comprise a substrate-binding region (yellow). Shown in red are the deduced CA-VA variants identified in this study. The index and affected brother of family 1 has a nonsynonymous Ser to Pro mutation at residue 233, adjacent to the substrate-binding region. The index of family 2 has a deletion of residues 154–186 (exon 4), thereby missing the metal-binding and active-site residues. The index of family 3 has a deletion of residues 207–258 (exon 6; the substrate-binding region), which results in absent protein.
