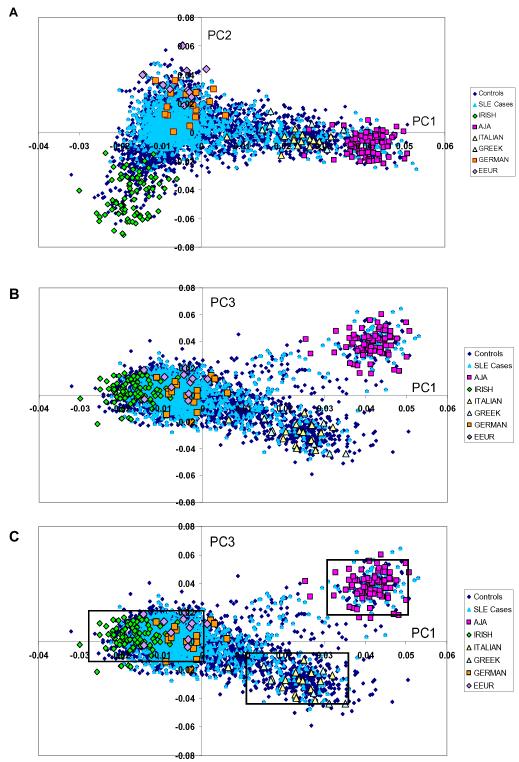
Figure 1.
Principal components analysis (PCA) shows European population substructure.
(a) Panel 1a plots principal component (PC) 2 against PC1. (b) Panel 1b plots PC3 against PC1. (c) Panel 1c identifies homogenous clusters (northern European, southern European, Ashkenazi Jewish) defined by PC1 and PC3. Points on the graphs represent individual study participants; individuals are color coded according to SLE status and grandparental country of origin, AJA=Ashkenazi Jewish ancestry, EEUR=Eastern European.
Population genetic substructure was determined using PCA of 4965 single nucleotide polymorphisms (SNPs) in EIGENSTRAT (Cambridge, MA). Of these SNPs, 2617 were informative for European population substructure and the remainder were informative for continental ancestry (187 SNPs) or East Asian substructure (2161 SNPs). These SNPs are distributed throughout the autosomal genome and exclude regions of extended linkage disequilibrium (inversion regions and the major histocompatibility complex region). Additionally, pairs of SNPs with r2 >0.5 in European population groups, SNPs not in Hardy-Weinberg equilibrium, and SNPs missing in >10% of participants were excluded.11
To improve PC differentiation and interpretation, we also included a set of 2398 controls genotyped for the same set of SNPs. Controls were adults of self-reported European ancestry enrolled in the New York Cancer Project who did not have SLE.19 Information on grandparental country of origin was collected for controls, and controls with all 4 grandparents from a single European country or region or who shared a single ethnic identity were used for PC interpretation.
Prior to PCA, we screened the study population for non-European ancestry using a set of 187 ancestry informative markers included in the SNP panel. We used STRUCTURE, a program that applies a model based, non-hierarchical clustering method for individual ancestry estimation.20 Participants with >10% non-European ancestry were excluded prior to PCA to determine European substructure. Participants were also excluded for outlying PC values (>6 standard deviations from the mean).