Abstract
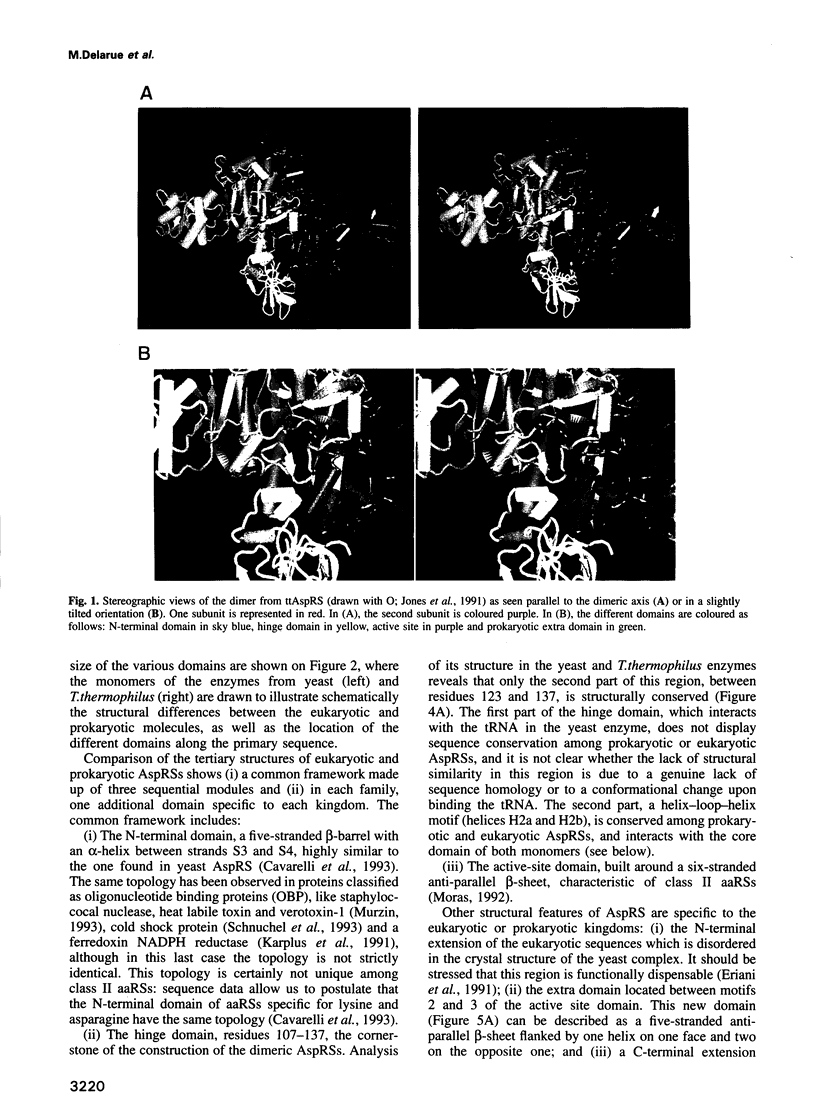
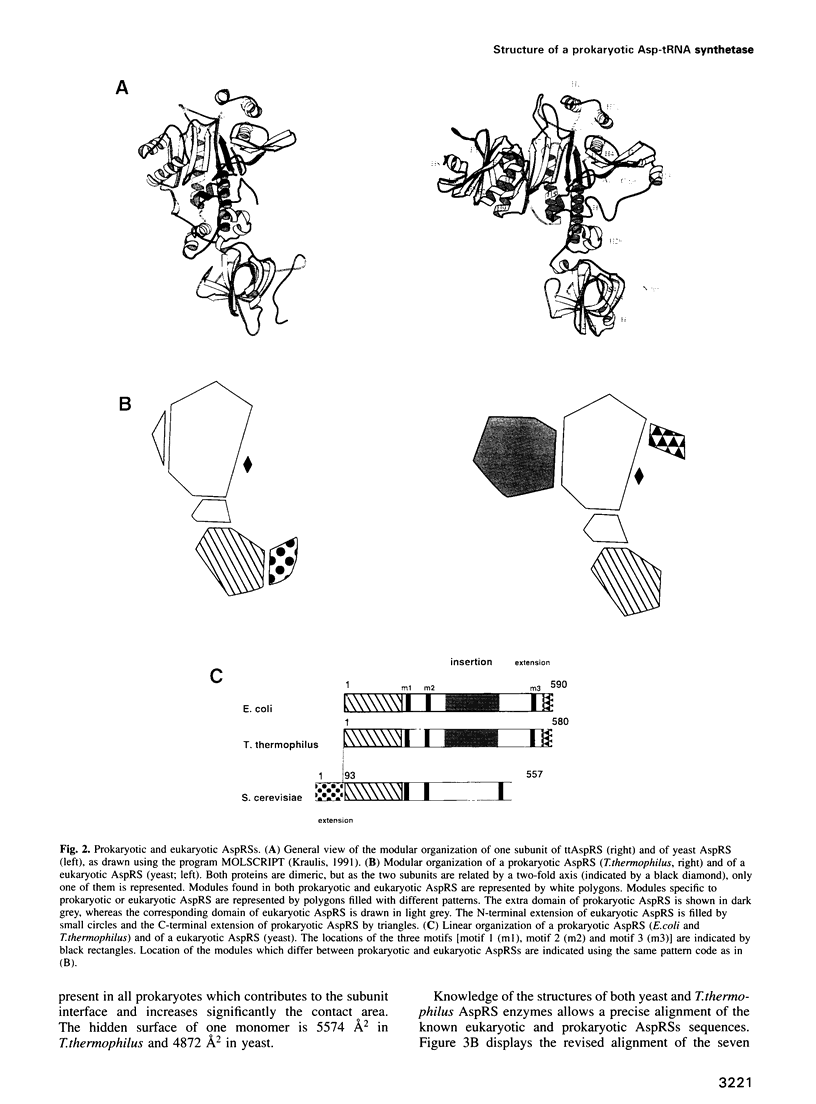
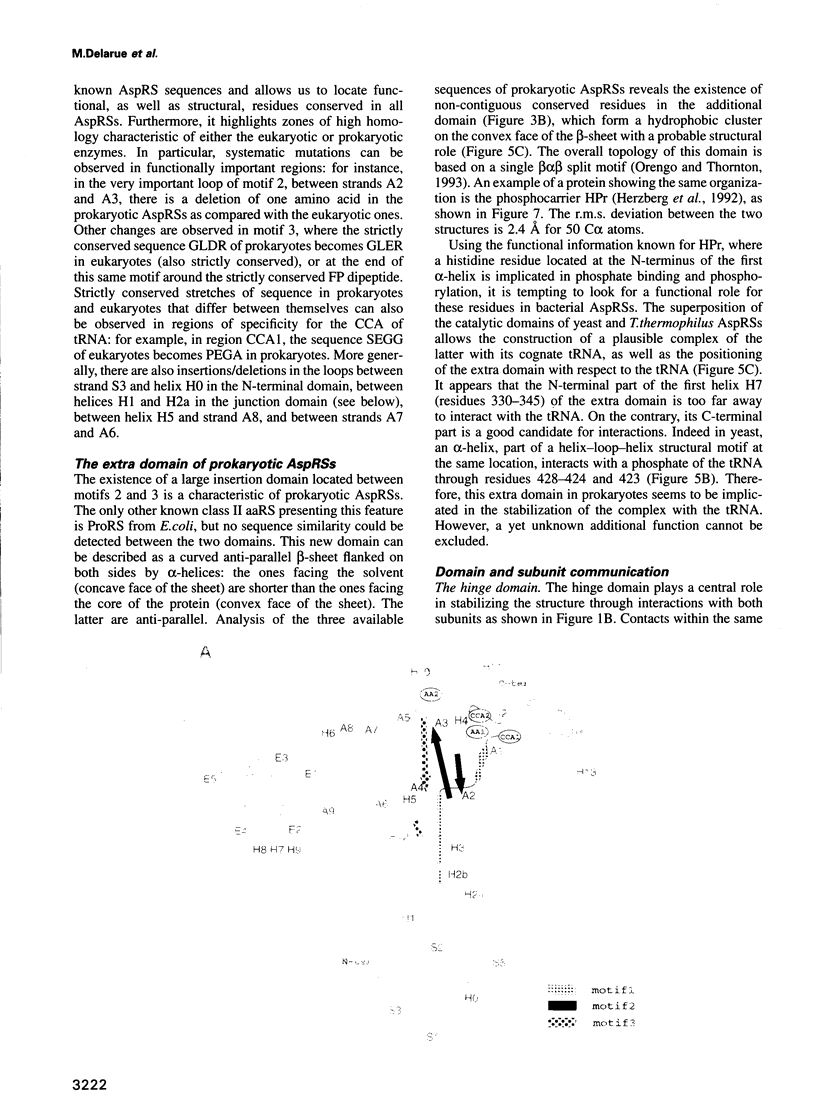
The crystal structure of Thermus thermophilus aspartyl tRNA-synthetase (AspRS) refined at 2.5 A resolution is described. This molecular structure is a textbook illustration of the modular organization of aminoacyl-tRNA synthetases. In addition to the three domains found in yeast AspRS, each monomer exhibits a module specific to prokaryotic enzymes, which corresponds to a helix-turn-helix motif in yeast AspRS, a domain implicated in the stabilization of the complex with tRNA. Its topology matches that of the histidine-containing phosphocarrier HPr which has been linked recently to another group of proteins containing the ferredoxin fold. We propose a more extensive alignment of these folds, which involves a circular permutation of the sequences and changes the point of entry of the whole domain. The C-terminal extension, another prokaryotic characteristic, leads to a significant increase in the network of interaction at the dimer interface. Some potential communication pathways suggest how a transfer of information between the two active sites of the homodimer might occur. Most of the residues involved belong to the class II-specific motifs in correlation with the dimeric state of nearly all class II enzymes. The T. thermophilus enzyme exhibits some features not found in any of the six other known AspRSs from mesophilic organisms.
Full text
PDF

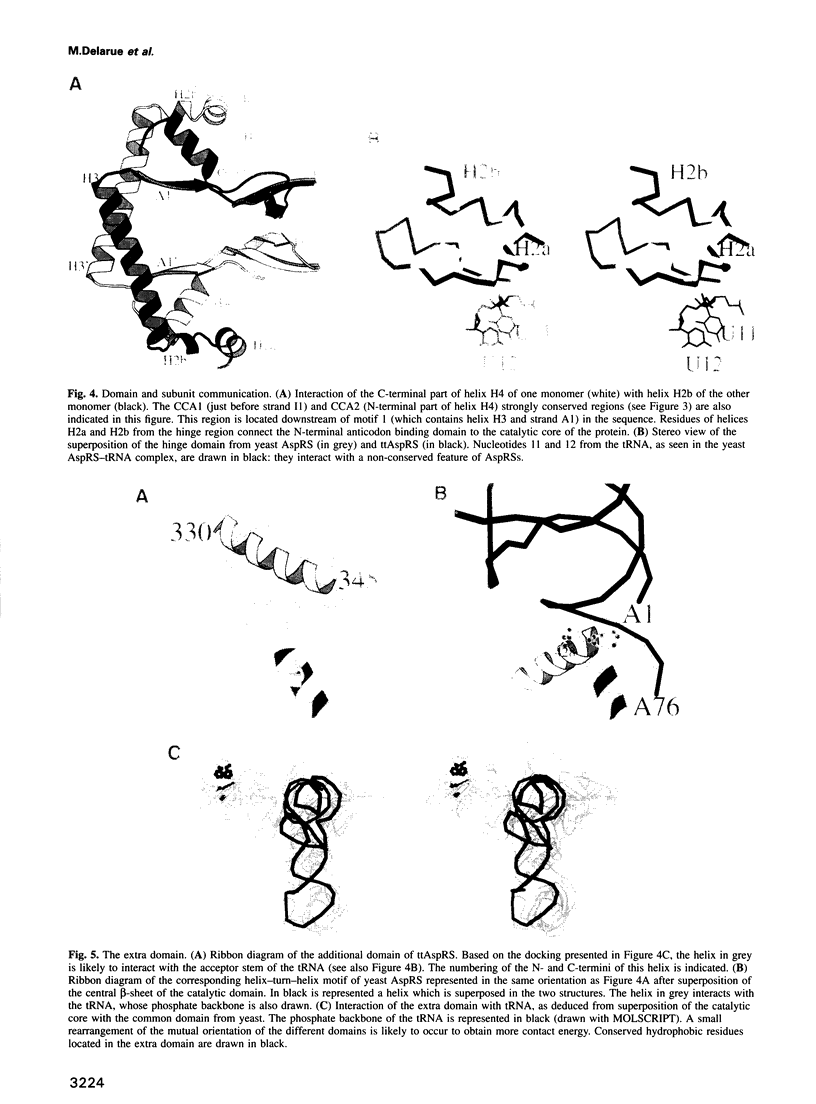
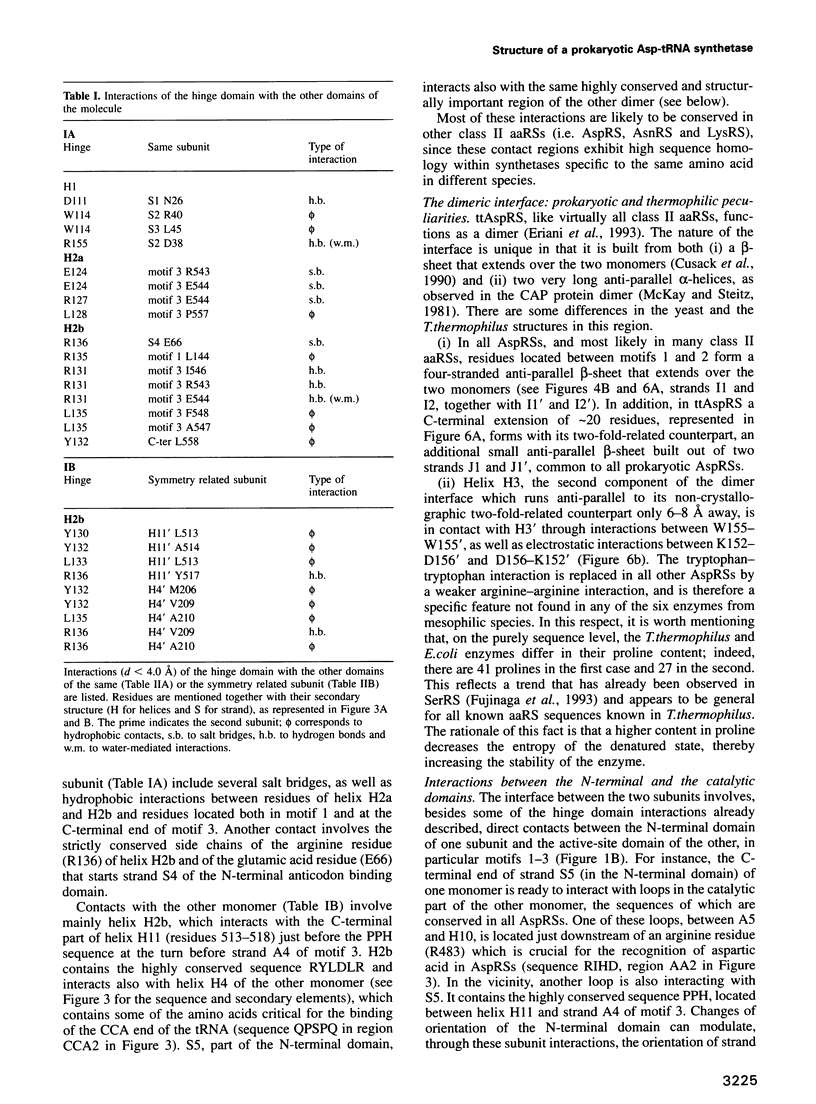
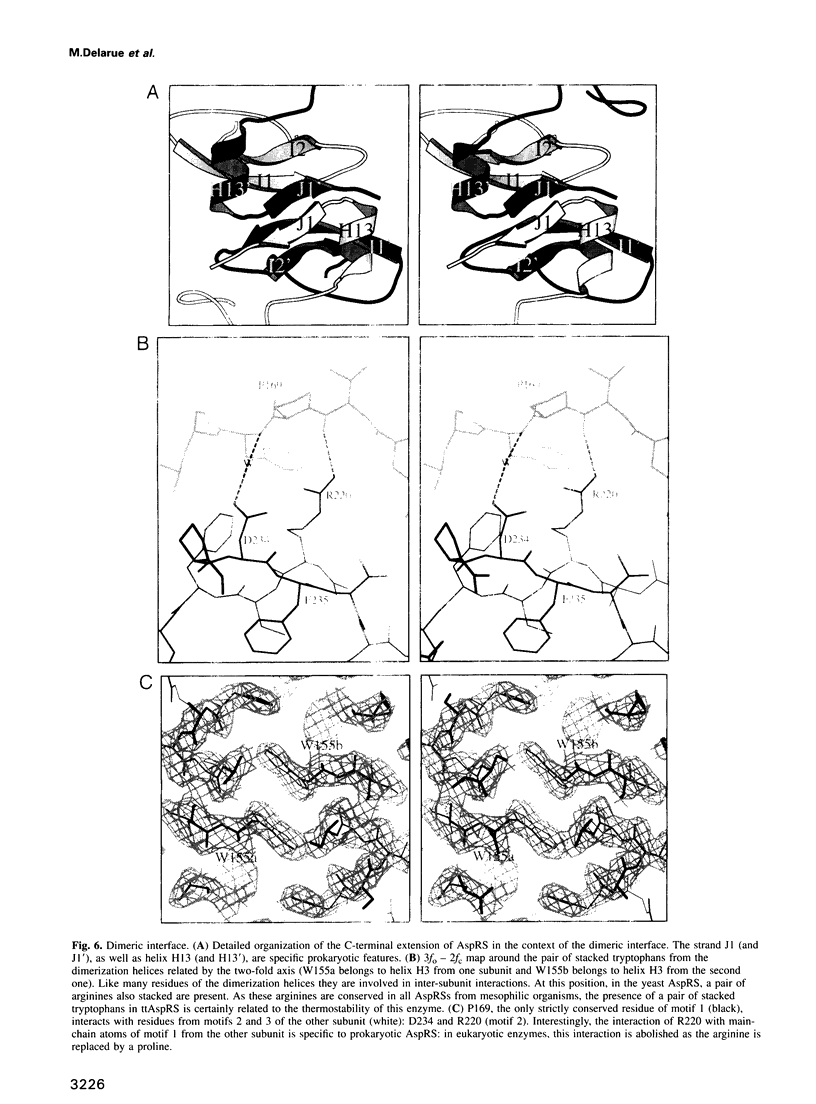
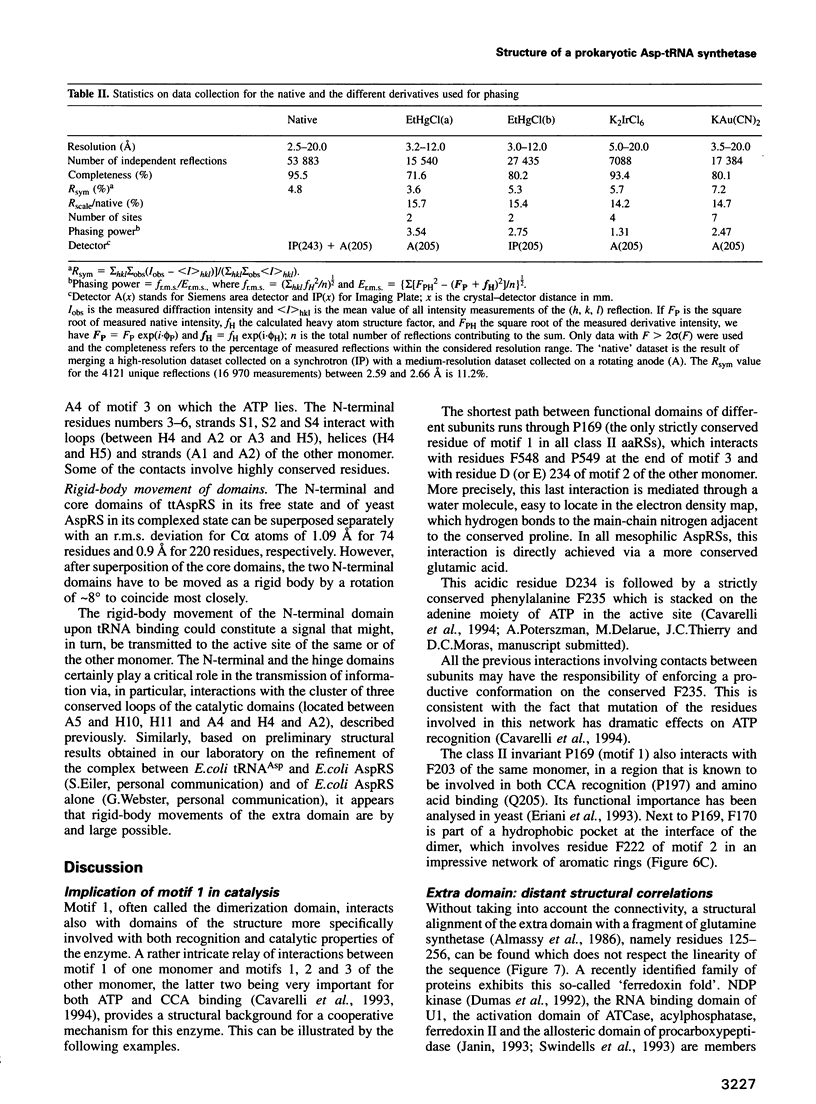


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almassy R. J., Janson C. A., Hamlin R., Xuong N. H., Eisenberg D. Novel subunit-subunit interactions in the structure of glutamine synthetase. 1986 Sep 25-Oct 1Nature. 323(6086):304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Eriani G., Rees B., Ruff M., Boeglin M., Mitschler A., Martin F., Gangloff J., Thierry J. C., Moras D. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994 Jan 15;13(2):327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Curmi P. M., Cascio D., Sweet R. M., Eisenberg D., Schreuder H. Crystal structure of the unactivated form of ribulose-1,5-bisphosphate carboxylase/oxygenase from tobacco refined at 2.0-A resolution. J Biol Chem. 1992 Aug 25;267(24):16980–16989. [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Delarue M., Moras D. The aminoacyl-tRNA synthetase family: modules at work. Bioessays. 1993 Oct;15(10):675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- Dumas C., Lascu I., Moréra S., Glaser P., Fourme R., Wallet V., Lacombe M. L., Véron M., Janin J. X-ray structure of nucleoside diphosphate kinase. EMBO J. 1992 Sep;11(9):3203–3208. doi: 10.1002/j.1460-2075.1992.tb05397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Cavarelli J., Martin F., Dirheimer G., Moras D., Gangloff J. Role of dimerization in yeast aspartyl-tRNA synthetase and importance of the class II invariant proline. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10816–10820. doi: 10.1073/pnas.90.22.10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Prevost G., Kern D., Vincendon P., Dirheimer G., Gangloff J. Cytoplasmic aspartyl-tRNA synthetase from Saccharomyces cerevisiae. Study of its functional organisation by deletion analysis. Eur J Biochem. 1991 Sep 1;200(2):337–343. doi: 10.1111/j.1432-1033.1991.tb16190.x. [DOI] [PubMed] [Google Scholar]
- Fujinaga M., Berthet-Colominas C., Yaremchuk A. D., Tukalo M. A., Cusack S. Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 A resolution. J Mol Biol. 1993 Nov 5;234(1):222–233. doi: 10.1006/jmbi.1993.1576. [DOI] [PubMed] [Google Scholar]
- Garber M. B., Yaremchuk A. D., Tukalo M. A., Egorova S. P., Fomenkova N. P., Nikonov S. V. Crystals of threonyl-tRNA synthetase from Thermus thermophilus. Preliminary crystallographic data. J Mol Biol. 1990 Aug 20;214(4):819–820. doi: 10.1016/0022-2836(90)90337-L. [DOI] [PubMed] [Google Scholar]
- Herzberg O., Reddy P., Sutrina S., Saier M. H., Jr, Reizer J., Kapadia G. Structure of the histidine-containing phosphocarrier protein HPr from Bacillus subtilis at 2.0-A resolution. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2499–2503. doi: 10.1073/pnas.89.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa N., Shimada N., Munakata Y., Watanabe K., Kimura N. Isolation and characterization of a gene encoding rat nucleoside diphosphate kinase. J Biol Chem. 1992 Jul 15;267(20):14366–14372. [PubMed] [Google Scholar]
- Janin J. Shared structural motif in proteins. Nature. 1993 Sep 2;365(6441):21–21. doi: 10.1038/365021a0. [DOI] [PubMed] [Google Scholar]
- Jasin M., Regan L., Schimmel P. Modular arrangement of functional domains along the sequence of an aminoacyl tRNA synthetase. Nature. 1983 Dec 1;306(5942):441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- Karplus P. A., Daniels M. J., Herriott J. R. Atomic structure of ferredoxin-NADP+ reductase: prototype for a structurally novel flavoenzyme family. Science. 1991 Jan 4;251(4989):60–66. [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Murzin A. G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993 Mar;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orengo C. A., Thornton J. M. Alpha plus beta folds revisited: some favoured motifs. Structure. 1993 Oct 15;1(2):105–120. doi: 10.1016/0969-2126(93)90026-d. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- Poterszman A., Plateau P., Moras D., Blanquet S., Mazauric M. H., Kreutzer R., Kern D. Sequence, overproduction and crystallization of aspartyl-tRNA synthetase from Thermus thermophilus. Implications for the structure of prokaryotic aspartyl-tRNA synthetases. FEBS Lett. 1993 Jul 5;325(3):183–186. doi: 10.1016/0014-5793(93)81069-c. [DOI] [PubMed] [Google Scholar]
- Rees B., Bilwes A., Samama J. P., Moras D. Cardiotoxin VII4 from Naja mossambica mossambica. The refined crystal structure. J Mol Biol. 1990 Jul 5;214(1):281–297. doi: 10.1016/0022-2836(90)90161-e. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Schnuchel A., Wiltscheck R., Czisch M., Herrler M., Willimsky G., Graumann P., Marahiel M. A., Holak T. A. Structure in solution of the major cold-shock protein from Bacillus subtilis. Nature. 1993 Jul 8;364(6433):169–171. doi: 10.1038/364169a0. [DOI] [PubMed] [Google Scholar]
- Swindells M. B., Orengo C. A., Jones D. T., Pearl L. H., Thornton J. M. Recurrence of a binding motif? Nature. 1993 Mar 25;362(6418):299–299. doi: 10.1038/362299a0. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990 Mar;8(1):52-6, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Wallace C. J. The curious case of protein splicing: mechanistic insights suggested by protein semisynthesis. Protein Sci. 1993 May;2(5):697–705. doi: 10.1002/pro.5560020501. [DOI] [PMC free article] [PubMed] [Google Scholar]