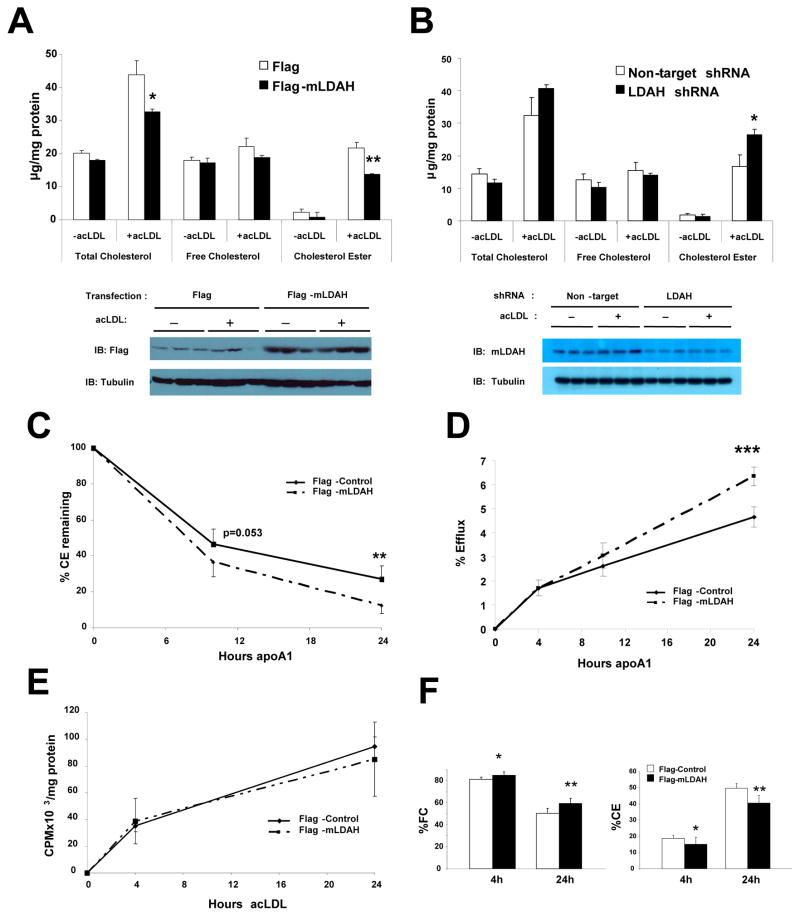
Figure 4. LDAH upregulation promotes cholesterol turnover from macrophages.
(A) RAW 264.7 macrophages were transfected with flag or flag-mLDAH plasmids. Cells were cultured for 18h in DMEM-1% FBS without or with acLDL (50 μg/ml). Cellular lipids were extracted, and total intracellular cholesterol, FC and CE levels were quantified and normalized to protein. (n= 3). Immunoblots on protein lysates of the cells used for the experiments are shown under the chart. (B) Total cholesterol, FC and CE levels in RAW 264.7 macrophages with lentiviral non-target shRNA or mLDAH shRNA-mediated knockdown. (n=3). Immunoblotts on protein lysates of RAW 264.7 macrophages with stable mLDAH knockdown are shown under the chart. (C) Time-course of CE hydrolysis in macrophages with and without mLDAH overexpression. RAW 264.7 macrophages were cholesterol-loaded with acLDL (50 μg/ml) for 20h in DMEM-0.2% BSA, washed, and pulsed with [3H] oleic acid (0.2 mM) in DMEM-0.4% BSA for 23h. Cells were then washed and pre-incubated with the ACAT1 inhibitor Sandoz 58–035 (10 μg/ml in DMEM-0.2% BSA) for 1h. ApoA1 (10 μg/ml) was added to the culture media that contained the ACAT1 inhibitor, and cells were harvested before and 6 and 24h after apoA1 was added. Intracellular lipids were resolved by TLC, and the CE at each timepoint was measured and normalized to protein. (n=6). (D) Effect of mLDAH overespression on cholesterol efflux. RAW 264.7 macrophages were loaded with [3H]-cholesterol-labeled acLDL (50 μg/ml in DMEM-0.2% BSA) for 18h, washed, equilibrated in DMEM-0.2% BSA for 1h, and cholesterol efflux was induced by adding apoA1 to the culture media. (n=5). (E and F) Time-course of cholesterol accumulation and distribution in the FC and CE fractions. RAW 264.7 macrophages were loaded with [3H]-cholesterol-labeled acLDL (50 μg/ml in DMEM-0.2% BSA) for 4 and 24h. Lipids were extracted and resolved by TLC, and FC and CE were quantified and normalized to protein. Total cholesterol (FC + CE) content is shown in panel (E), and the cholesterol distribution in the FC and CE fractions is shown in panel (F). (n=5). All data are shown as mean ± SD. *p<0.05, **p<0.01, ***p<0.001.