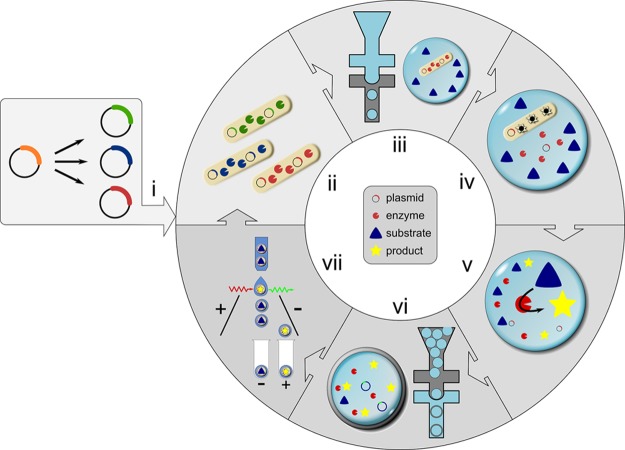
Figure 1.
Generating double emulsions on two chips and selection of active biocatalysts. The workflow for one cycle of directed evolution consists of the following steps: (i) Gene libraries are generated from an enzyme-encoding plasmid. (ii) E. coli cells produce the biocatalyst of interest in liquid culture. (iii) In a first microfluidic device (with hydrophobic, fluorocarbon-coated channel walls), single cells are compartmentalized in droplets together with substrate and lysis agents. (iv) After cell lysis, substrate and cytoplasmically expressed enzyme react to yield a fluorescent product. (v) The reaction is allowed to proceed for a desired incubation period (in our case up to 24 h, but droplets are stable for at least one month). The reaction progress can be stopped simultaneously in all water-in-oil droplets by heat inactivation, so that the time required for double emulsion formation and sorting does not extend the assay period. (vi) Next, primary droplets are transformed into double emulsions in a second device with identical design to the one used in (iii) but with hydrophilic coating. (vii) Variants exhibiting the highest activity are identified and sorted in a standard flow cytometer. The recovered DNA can be used for further rounds of evolution without PCR amplification when a high-copy plasmid is used. The procedure takes little time: droplet formation (steps iii and vi) takes place at a frequency of 6–12 kHz, so that a library of 107 double emulsion droplets is produced in 90 min. Sorting 107 droplets at a rate of 10–15 kHz takes about 15 min.