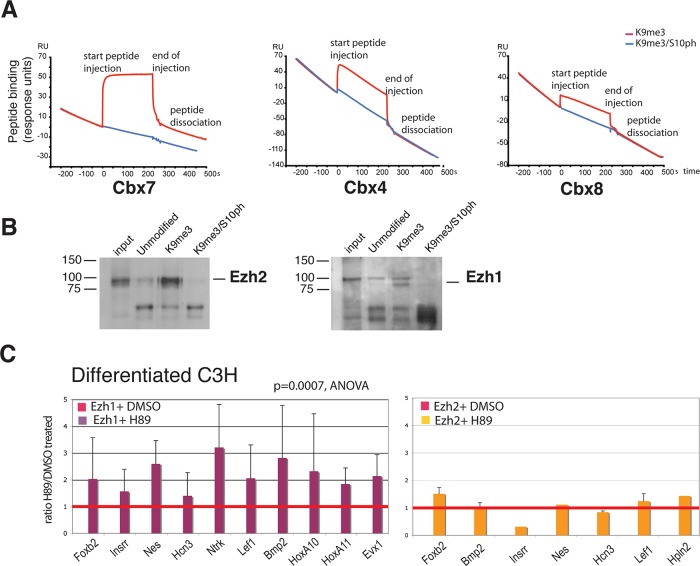
FIGURE 6:
Binding of Polycomb proteins to H3K9me3 is affected by the H3S10ph modification. (A) SPR was used to measure binding of purified histidine (His)-tagged Cbx4, 7, and 8 to peptides corresponding to amino acids 1–15-Y of histone H3 containing the K9me3 or the K9me3/S10ph modifications. Peptides were injected over the indicated His-Cbx proteins immobilized on a Biacore flow cell (see Materials and Methods). Red and blue lines indicate binding of K9me3 and K9me3/S10ph peptides, respectively. (B) Binding of Ezh1- and Ezh2-containing PRC2 complexes to histone H3 peptides was assessed by peptide capture. Nuclear extracts from NIH3T3 fibroblasts were used for peptide capture assays as described in the Supplemental Methods. Pull downs were performed with biotinylated peptides derived from the N-terminus of histone H3, corresponding to amino acids 1–20. The peptides were unmodified or contained K9me3 or combined K9me3/S10ph. Captured proteins were identified by Western blot. (C) Reduction of the H3K9me3/S10ph modification by treatment with H89 increases recruitment of Ezh1 to Polycomb-repressed genes but has little effect on Ezh2 binding. Binding of Ezh1 and Ezh2 to promoters of repressed genes was analyzed after treatment with H89 or vehicle (dimethyl sulfoxide). The effect of H89 on the levels of Ezh1 binding across a panel of 13 genes was statistically significant (p = 0.0007, analysis of variance). Bars show mean ± SD. n = 7 biological replicates. No significant effect of H89 treatment was observed on binding of Ezh2 to repressed genes, consistent with its absence from silent genes in these cells.