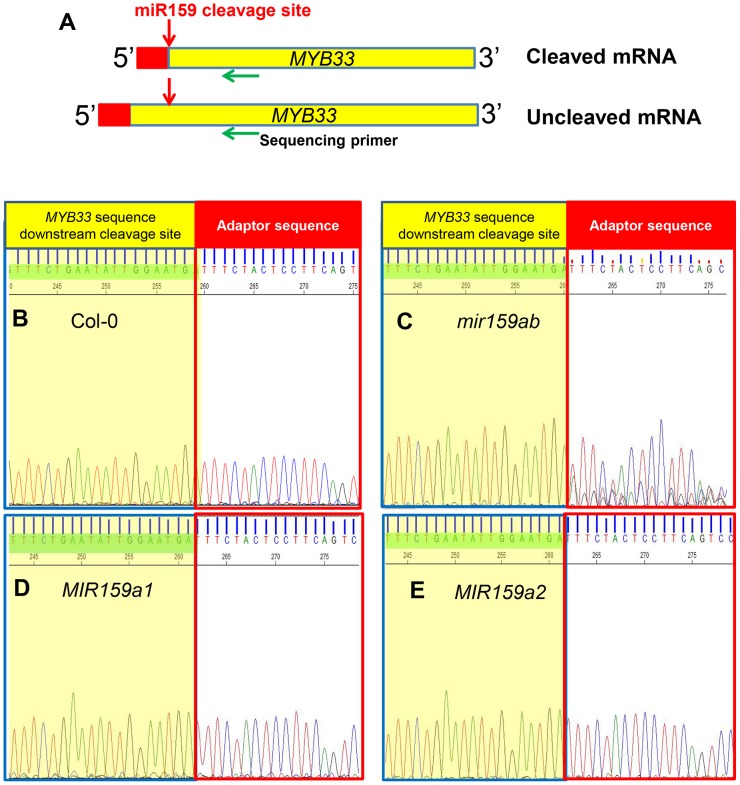
Figure 3. MiR159a variants with up to two central mismatches can direct target cleavage at the canonical miR159 cleavage site.
(A) Schematic representation of MYB33 PCR products after a modified 5′-RACE procedure to determine the proportion of degraded MYB33 mRNA that corresponds to miR159-guided cleavage products. Red box: RNA Oligo adaptor; Yellow box: MYB33 sequences; Red arrow: canonical miR159 cleavage site; Green arrow: MYB33 sequencing primer. (B–E) Sequencing chromatographs of the cDNA-adaptor region from 5′-RACE recovered 3′ MYB33 transcripts in: (B) Col-0, (C) mir159ab, (D) MIR159a1 and (E) MIR159a2 plants. Peaks from MYB33 sequence downstream of the miR159 cleavage sites are highlighted in yellow, while those from the adaptor are left white. Three independent transgenic lines were checked for each construct and the results were identical.