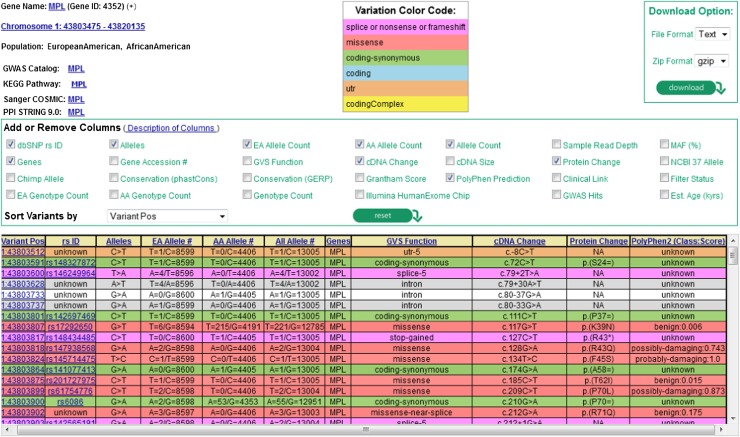
Figure 4.
Screenshot from exome variant server of the MPL gene showing part of a summary table of variants discovered through the NHLBI ESP. The current data release is taken from 6503 unrelated European American and African American samples drawn from multiple ESP cohorts and represents all of the ESP exome variant data. Users can select summary characteristics of interest for display and query sequence variants by gene, rsID (the variant identifier in dbSNP, if known), chromosomal location, or batch. The corresponding attributes (eg, allele counts or frequencies overall or by ethnicity, various evolutionary conservation scores such as GERP, phastCons, functional annotation) can be viewed on the web or downloaded as text-formatted files. Color coding is used to annotate variants according to genomic function (eg, splice/nonsense/frameshift, missense, synonymous, UTR). A functional prediction for each missense variant is shown using the Polyphen2 prediction algorithm.86 GERP, genomic evolutionary rate profiling; UTR, untranslated region.