FIGURE 6.
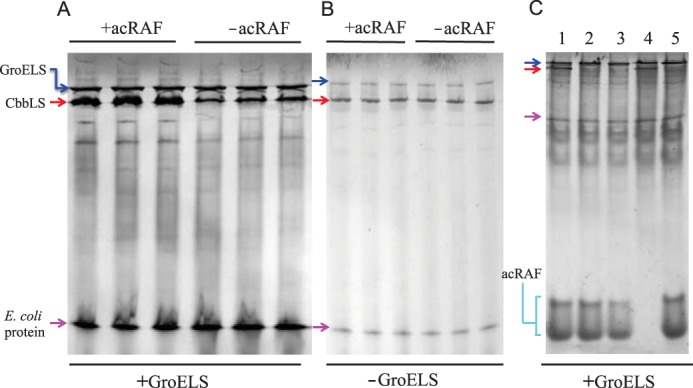
acRAF increases the quantity of natively assembled RuBisCO when heterologously co-expressed with GroELS in E. coli. A, the RuBisCO CbbLS genes were co-overexpressed with GroELS in E. coli BL21(DE3) either with (+) or without (−) acRAF. Soluble, whole cell lysates were analyzed by native non-denaturing PAGE. Independent co-expression experiments were performed in triplicate. The amount of natively assembled RuBisCO is substantially increased by the presence of acRAF (when GroELS is also overexpressed). The band corresponding to assembled CbbLS was deduced by comparisons to control gels run on lysates from cells expressing or lacking specific proteins (panel C). B, CbbLS was overexpressed in E. coli BL21(DE3) either with (+) or without (−) acRAF, but without GroELS overexpression. Soluble, whole cell lysates were analyzed by native non-denaturing PAGE. Independent co-expression experiments were performed in triplicate. No effect of acRAF on RuBisCO production is seen. C, CbbLS and acRAF bands were identified from soluble fractions of lysates of E. coli BL21(DE3) cells, which were transformed with different combinations of compatible plasmids. Strain 1, CbbLS, acRAF and GroELS. Strain 2, CbbL, acRAF, GroELS. Strain 3, CbbS, acRAF, GroELS. Strain 4, CbbLS, empty pET22b, GroELS. Strain 5, empty pRSF, acRAF, GroELS. RuBisCO (hexadecameric L8S8) and acRAF band identities were deduced by comparisons of strain-specific band patterns. The acRAF from H. neapolitanus migrated as two species, in contrast to acRAF from T. intermedia, which migrates as one species (not shown). Note that only strains 1 and 4 are expressing both large and small RuBisCO subunits, consistent with the assignment of native RuBisCO to the band present exclusively in those lanes. Blue, red, and magenta arrows indicated bands that correspond to GroELS, CbbLS, and an unidentified E. coli protein, respectively.
