Abstract
Allosteric modulators of A1 and A2A adenosine receptors have been described; however, for the A3 adenosine receptor, neither an allosteric site nor a compound with allosteric effects has been described. In this study, the allosteric modulation of human A3 adenosine receptors by a series of 3-(2-pyridinyl)isoquinoline derivatives was investigated by examining their effects on the dissociation of the agonist radioligand, [125I]N6-(4-amino-3-iodobenzyl)-5′ -N-methylcarboxamidoadenosine (I-AB-MECA), from the receptor. Several 3-(2-pyridinyl)isoquinoline derivatives, including VUF5455, VUF8502, VUF8504, and VUF8507, slowed the dissociation of the agonist radioligand [125I]I-AB-MECA in a concentration-dependent manner, suggesting an allosteric interaction. These compounds had no effect on the dissociation of the radiolabeled antagonist [3H]PSB-11 from the A3 adenosine receptor, suggesting a selective enhancement of agonist binding. By comparison, compounds of similar structure (VUF8501, VUF8503, VUF8505), the classical adenosine receptor antagonist CGS15943 and the A1 receptor allosteric enhancer PD81723 did not significantly influence the dissociation rate of [125I]I-AB-MECA. The effect of agonist on forskolin-induced cAMP production was significantly enhanced by VUF5455. When the subtype-selectivity of the allosteric enhancement was tested the compounds had no effect on the dissociation of either [3H]N6-[(R)-phenylisopropyl]adenosine from the A1 adenosine receptor or [3H]CGS21680 from the A2A adenosine receptor. Probing of structure-activity relationships suggested that a carbonyl group is essential for allosterism but preferred only for competitive antagonism. The presence of a 7-methyl group decreased the competitive binding affinity without a major loss of the allosteric enhancing activity, suggesting that the structural requirements for allosteric enhancement might be distinct from those for competitive antagonism.
The purine nucleoside adenosine produces numerous physiological actions via cell surface adenosine receptors. These receptors are widely distributed throughout the body and are subclassified as A1, A2A, A2B, and A3 adenosine receptors (Fredholm et al., 2000). All of the receptors belong to the G protein-coupled receptor (GPCR) family.
The A3 adenosine receptor is the adenosine receptor subtype identified most recently (Zhou et al., 1992) and is involved in a variety of physiological processes (Kaiser and Quinn, 1999). Stimulation of the A3 adenosine receptor increases the release of inflammatory mediators, such as histamine, from mast cells (Hannon et al., 1995). Tumor necrosis factor-α production is inhibited at the level of transcription by the activation of the A3 receptor, which suppresses steady-state mRNA levels (Sajjadi et al., 1996). The A3 adenosine receptor is believed to be involved in ischemic preconditioning of the heart and kidney (Strickler et al., 1996; Lee and Emala, 2000). The activation of the A3 adenosine receptor is also suggested to be involved in immunosuppression (MacKenzie et al., 1994) and brain ischemia (Von Lubitz et al., 1994). Thus, adenosine receptor agonists and agents that enhance the response to adenosine would have many clinic applications. The clinical application of adenosine analogs as directly acting, potent adenosine receptor agonists has not yet proven successful because of potential side effects. Side effects are caused by the ubiquitous presence of adenosine receptors throughout the body, which are activated by agonists indiscriminately. Thus, it is useful to consider an alternative approach that may activate adenosine receptors in a more specific manner.
Several members of the GPCR superfamily have been reported to be modulated allosterically (Birdsall et al., 1995). Allosteric modulation of GPCRs has been characterized most extensively for muscarinic receptors (Holzgrabe and Mohr, 1998), and it has been suggested that allosteric modulators may provide therapeutic advantages over orthosteric agonists. Such advantages may include greater subtype selectivity and fewer side effects (Birdsall et al., 1995; Bhattacharya and Linden, 1996; Linden, 1997). For example, diazepam and other benzodiazepines, which act as allosteric enhancers of the ion channel-coupled GABAA receptor, have acceptable side effects and are used clinically. In contrast, directly acting GABAA agonists have widespread side effects and are not used clinically. The effects of an allosteric enhancer on an organ or tissue might be event-specific because of an increase in the local concentration of the endogenous agonist. For example, hypoxic conditions increase the local production of cytoprotective adenosine. Compounds that either augment the concentration of adenosine or enhance its action locally may have a better therapeutic profile than the agonists. Additionally, neurotransmitter receptors have been reported to be less sensitive to desensitization or down-regulation by allosteric enhancers than by exogenous agonists (Birdsall et al., 1995). Thus, allosteric modulators could offer a control of receptor function not found with competitive agonists.
Within a family of receptors that bind the same endogenous ligand, the primary binding sites may be similar, because the amino acids that form this site are highly conserved (Tucek and Proska, 1995). An allosteric site, being spatially distinct from the primary site, may be located at less conserved regions on the receptor, which may provide greater subtype selectivity (Tucek and Proska, 1995; Birdsall et al., 2001).
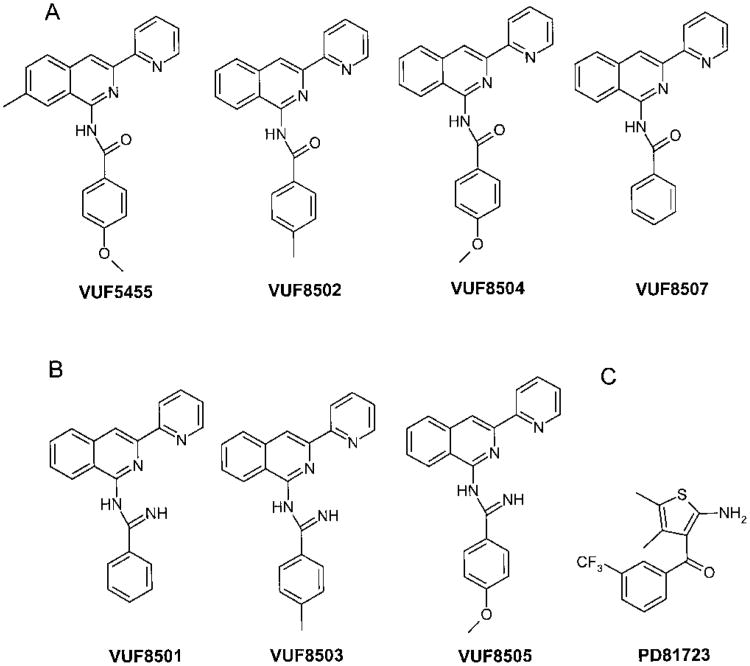
Allosteric modulation of A1 adenosine receptors was reported (Bruns and Fergus, 1990; Linden, 1997). A number of aminobenzoylthiophenes, including PD81723, were allosteric modulators of the A1 adenosine receptor (Bruns and Fergus, 1990). These compounds were shown to be highly subtype-selective enhancers for A1 adenosine receptors (Bruns and Fergus, 1990) and were less likely to cause desensitization and down-regulation of receptors than selective A1 adenosine receptor agonists (Bhattacharya and Linden, 1996). Recently, an allosteric site on the A2A adenosine receptor was defined (Gao and IJzerman, 2000; Gao et al., 2000). However, it was not known whether there is an allosteric binding site on the A3 adenosine receptor; compounds with allosteric effects at A3 adenosine receptors have not been reported previously. This study demonstrated allosteric modulation of A3 adenosine receptors by a number of 3-(2-pyridinyl)isoquinoline derivatives (Fig. 1, A and B).
Fig. 1.
Chemical structures of the compounds tested in this study. A, isoquinoline derivatives found to enhance A3 agonist binding. B, isoquinoline derivatives that did not enhance A3 agonist binding. C, A1 allosteric enhancer.
Experimental Procedures
Materials
3-(2-Pyridinyl)isoquinoline derivatives were synthesized at Leiden/Amsterdam Center for Drug Research (Amsterdam, The Netherlands). [125I]N6-(4-amino-3-iodobenzyl)adenosine-5′-N-methyluronamide ([125I]I-AB-MECA; 2000 Ci/mmol), [3H]PSB-11 (53 Ci/mmol), [3H]N6-[(R)-phenylisopropyl]adenosine ([3H]R-PIA; 34 Ci/mmol), and [3H]CGS21680 (47 Ci/mmol) were from Amersham Pharmacia Biotech (Buckinghamshire, UK). Adenosine deaminase was obtained from Roche Molecular Biochemicals (Mannheim, Germany). Bicinchoninic acid (BCA) and BCA protein assay reagent were from Pierce Chemical (Rockford, IL). CGS15943 was a gift from Novartis (Summit, NJ). CPA and NECA were obtained from Sigma/RBI (Natick, MA). Rolipram and forskolin were purchased from Sigma (St. Louis, MO). cAMP (low pH) Immunoassay kits were from R & D Systems (Minneapolis, MN).
Membrane Preparation
Forebrain and striatal tissue from Wistar rats were homogenized in ice-cold 50 mM Tris-HCl buffer, pH 7.7, using an electric homogenizer. The homogenate was centrifuged at 50,000g for 10 min at 4°C, and the pellet was washed in fresh buffer. A pretreatment with adenosine deaminase (2 units/ml) was performed. The final pellet was stored at −80° until the binding experiments. HEK293 cells expressing human A3 adenosine receptors and native RBL-2H3 mast cells with native rat A3 adenosine receptors were harvested by trypsinization. Cells were centrifuged at 500g for 10 min, and the pellet was suspended in 50 mM Tris-HCl buffer, pH 8.0, containing 10 mM MgCl2, 1 mM EDTA and 0.1 mg/ml CHAPS. The suspension was homogenized with an electric homogenizer for 5 s, and was then recentrifuged at 18,000g for 30 min at 4°C. The resultant pellets were resuspended in buffer in the presence of 2 U/ml adenosine deaminase, and the suspension was stored at −80°C. Protein concentrations were measured with the BCA method (Smith et al., 1985).
Competitive Binding Assays
Binding of 0.15 nM [125I]I-AB-MECA (Olah et al., 1994) to membranes of HEK293 cells expressing human A3 adenosine receptors (25 μg of protein) was carried out in duplicate at 37°C for 1 h in 100 μl of 50 mM Tris-HCl buffer, pH 8.0, containing 10 mM MgCl2, 1 mM EDTA, and 0.1 mg/ml CHAPS. Nonspecific binding was defined as binding in the presence of 30 μM NECA. Tested compounds were dissolved in dimethyl sulfoxide. Control incubations also contained the same concentrations of dimethyl sulfoxide (≤ 1%). The reaction was terminated by filtration through GF/B filters on a cell harvester (Brandel, Gaithersburgh, MD).
Dissociation of the Radiolabeled Adenosine Receptor Agonist, [125I]I-AB-MECA, from Human and Rat A3 Adenosine Receptors
[125I]I-AB-MECA (0.15 nM) was preincubated with the A3 adenosine receptor membranes at 37°C for 1 h. Nonspecific binding was determined in parallel by addition of 30 μM NECA before the preincubation. Dissociation was then initiated by the addition of NECA (final concentration, 30 μM) and test agents. Nonspecific binding in the presence of tested agents was also determined in parallel. After an additional 1.5 h at 37°C, the dissociation was terminated by filtration through GF/B filters on a cell harvester.
Dissociation of the Radiolabeled, Selective Antagonist [3H]PSB-11 from A3 Adenosine Receptors
Membranes (60 μg) were preincubated with 4 nM [3H]PSB-11 (selective A3 receptor antagonist; Müller, 2001) in a total assay volume of 100 μl for 120 min. Dissociation was initiated by addition of 30 μM NECA with or without test compounds. The time course of dissociation was measured by rapid filtration at appropriate time intervals. Nonspecific binding was measured after a 120-min incubation in the presence of 30 μM NECA.
Dissociation of [3H]R-PIA from A1 Adenosine Receptors
Binding of 1 nM [3H]R-PIA to A1 adenosine receptors in rat forebrain membranes was carried out at 37°C for 90 min in 50 mM Tris-HCl buffer, pH 7.7, with a total assay volume of 400 μl. Dissociation was initiated by the addition of 10 μM CPA with or without test compounds. Nonspecific binding was determined using 10 μM CPA. Samples were filtered after incubation at 37°C at the various time points.
Dissociation of [3H]CGS21680 from A2A Adenosine Receptors
Rat striatal membranes (80 μg) were incubated with 15 nM [3H]CGS21680 at 25°C for 90 min in 400 μl of 50 mM Tris-HCl, pH 7.7, containing 10 mM MgCl2. NECA (10 μM) was used to define nonspecific binding. Dissociation was initiated by the addition of 10 μM NECA in the presence or absence of tested compounds.
Measurement of cAMP Level
cAMP production was measured by using a commercially available, low-pH cAMP Immunoassay Kit (R&D Systems, Inc.). Briefly, HEK293 cells expressing recombinant human A3 adenosine receptors were grown to 70% confluence in 12 well plates and then treated with test compounds. Thirty minutes after the treatment, 10 μM forskolin was added to the culture medium to stimulate cAMP levels, and was incubated for an additional 15 min at 37°C. Reaction was terminated by the addition of 1 ml of 0.1 N HCl, and the cellular debris was removed by centrifugation for 5 min at 10,000g. All experiments were performed in the presence of 10 μM rolipram and 3 U/ml adenosine deaminase. cAMP levels were measured using a Bio-kinetics reader (Bio-Tek instruments Inc., Winooski, VT).
Data Analysis
Binding experiment parameters were estimated using Prism software (GraphPAD, San Diego, CA). IC50 values obtained from competition curves were converted to Ki values by using the Cheng-Prusoff equation.
Results
Effects of the Test Compounds on the Dissociation of [125I]I-AB-MECA and [3H]PSB-11 from Human A3 Adenosine Receptors Expressed in HEK293 Cells
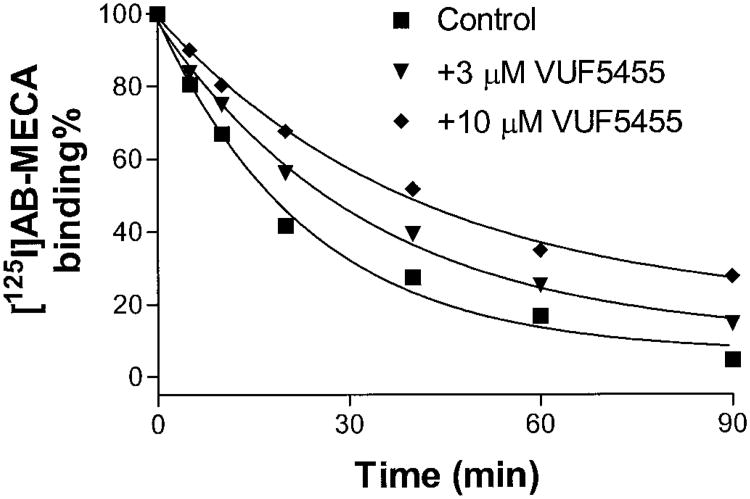
The dissociation of [125I]I-AB-MECA from A3 adenosine receptors following the addition of 30 μM NECA (Fig. 2) was characterized in the absence or presence of the test compounds. The dissociation rate (k−1) in the absence of the test compounds was 0.042 ± 0.005 min−1, and the t1/2 of dissociation was 16.4 ± 2.5 min. The k−1 values and the half-life in the presence of 3 μM (k−1 = 0.031 ± 0.003 min−1; t1/2 = 23.2 ± 3.4 min) and 10 μM (k−1 = 0.024 ± 0.003 min−1; half-life = 28.5 ± 4.3 min) VUF5455 (Fig. 1A) were significantly different from those of the control values (p < 0.05). Compounds VUF8502, VUF8504 and VUF8507 also significantly decreased the dissociation at 10 μM (Table 1). The potencies of these compounds, as determined by their ability to influence the dissociation rate, were very similar. By contrast, the isoquinoline derivatives VUF8501, VUF8503, and VUF8505 (Fig. 1B), the A1 allosteric enhancer PD81723 (Fig. 1C) and the classical nonselective adenosine receptor antagonist CGS15943 did not influence the dissociation rate significantly (Table 1), even at 30 μM (data not shown). The k−1 values in the presence or absence of the test compounds (10 μM) are listed in Table 1. The kinetics of [125I]I-AB-MECA association to the A3 receptor was also determined, and the association rate constant k1 was calculated to be 0.061 ± 0.022 min−1 nM−1. The kinetic Kd value calculated by using k1 and k−1 was 0.69 ± 0.23 nM.
Fig. 2.
Effects of VUF5455 on the dissociation of [125I]I-AB-MECA from human A3 adenosine receptors expressed in HEK293 cells. [125I]I-AB-MECA (0.15 nM) was preassociated with the A3 adenosine receptor membranes for 1 h at 37°C in a total assay volume of 100 μl. Nonspecific binding was determined in parallel by addition of 30 μM NECA before the preassociation. After 1 h preassociation, dissociation was initiated by addition of NECA (final concentration, 30 μM) alone or with VUF5455. The incubations were terminated by filtration through GF/B filters on a cell harvester (Brandel) after additional times as indicated. The data shown were derived from one experiment performed in duplicate and are typical of three independent experiments giving similar results.
Table 1.
The Ki values of tested compounds for [125I]I-AB-MECA (0.15 nM) binding to human A3 adenosine receptors and the dissociation rates (k−1) in the absence and presence of the tested compounds (10 μM). The assay was carried out as described under Experimental Procedures. Data shown are the mean ±S.E. from three independent experiments performed in duplicate.
| Ki (nM) | k−1 (min −1) | |
|---|---|---|
| nM | ||
| Control | 0.042 ± 0.007 | |
| VUF8501 | 739 ± 146 | 0.043 ± 0.004 |
| VUF8502 | 96 ± 26 | 0.024 ± 0.003* |
| VUF8503 | 581 ± 82 | 0.041 ± 0.011 |
| VUF8504 | 17.3 ± 1.7 | 0.022 ± 0.004* |
| VUF8505 | 307 ± 99 | 0.044 ± 0.012 |
| VUF8507 | 204 ± 42 | 0.021 ± 0.002* |
| VUF5455 | 1680 ± 620 | 0.024 ± 0.003* |
| PD81723 | N.D. | 0.046 ± 0.009 |
| CGS15943 | 185 ± 37 | 0.045 ± 0.005 |
N.D., not determined.
p < 0.05 compared with control (Student's t test).
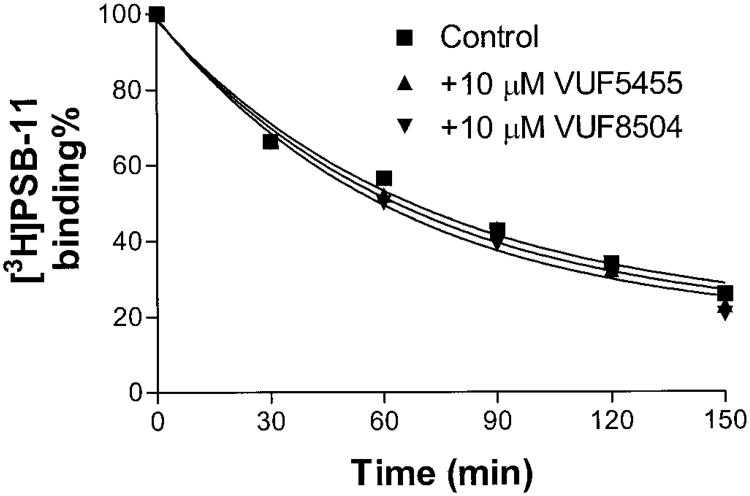
The dissociation rate of the novel A3 selective antagonist [3H]PSB-11 from A3 adenosine receptors expressed in HEK293 cells was either unaffected or slightly increased by these compounds (Fig. 3), suggesting that slowing of the dissociation was specific for the agonist state of the A3 adenosine receptor.
Fig. 3.
Time course of the dissociation of [3H]PSB-11 from the human A3 adenosine receptor. Membranes (60 μg) were preincubated with 4 nM [3H]PSB11 in a total assay volume of 100 μl for 120 min at 4°C. The dissociation of [3H]PSB-11 from the A3 adenosine receptor was started by addition of 30 μM NECA in the absence or presence of tested compounds. Nonspecific binding was measured after 120 min incubation in the presence of 30 μM NECA. The reaction was measured by rapid filtration at appropriate time intervals. Data are from one experiment performed in duplicate; two additional experiments gave similar results.
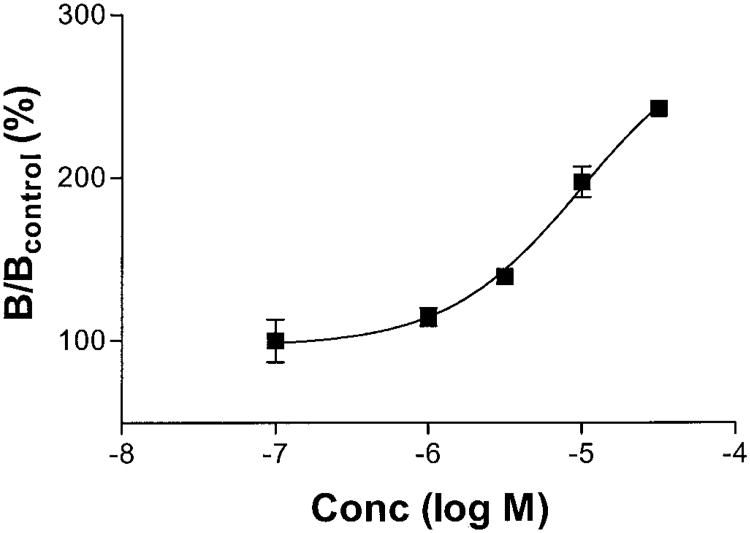
Concentration-Response Relationship for the Slowing of [125I]I-AB-MECA Dissociation from Human A3 Adenosine Receptors by Allosteric Enhancers
Fig. 4 shows the influence of increasing concentrations of the test compounds on the dissociation of [125I]I-AB-MECA in the presence of 30 μM NECA. Dissociation was allowed to proceed for 60 min before the reaction was terminated by filtration. Low concentrations of VUF5455 (<1 μM) did not modify the dissociation of [125I]I-AB-MECA. However, at concentrations > 1 μM, the dissociation rate decreased significantly. Because of the limited solubility of these compounds, the highest concentration used was 30 μM. It seemed that higher concentrations of these compounds produced more pronounced effects.
Fig. 4.
Concentration-response curve for the slowing of the dissociation of [125]I-AB-MECA by VUF5455. [125I]I-AB-MECA (0.15 nM) was preassociated with human A3 receptors in HEK293 cell membranes (25 μg protein) for 60 min at 37°C without additions (total binding) or in the presence of 30 μM NECA (nonspecific binding). At the end of the incubation period, 30 μM NECA was added simultaneously with vehicle or various concentrations of the tested compounds. The incubation was terminated after an additional 60 min. Control specific binding (Bcontrol) at the end of the 60-min dissociation period was approximately 500 cpm, approximately 20% of the total binding. Data are mean values from three independent experiments performed in duplicate.
Competition of Test Compounds with [125I]I-AB-MECA Binding for Human A3 Adenosine Receptors Expressed in HEK293 Cells
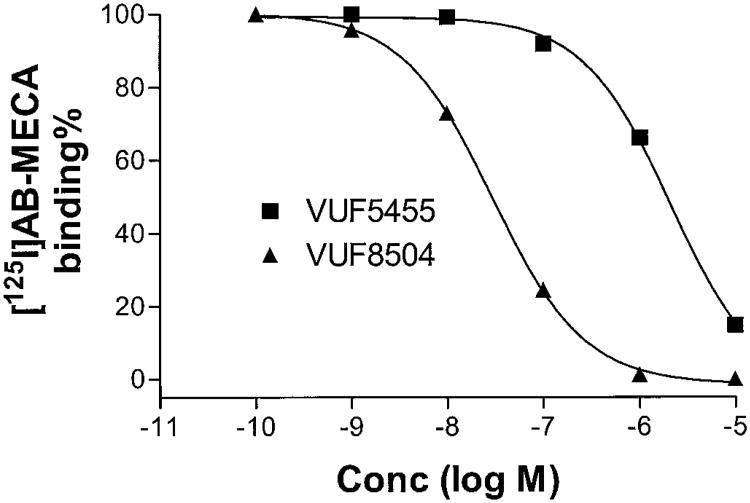
Fig. 5 shows the competition of [125I]I-AB-MECA binding to A3 adenosine receptors expressed in HEK293 cells by the 3-(2-pyridinyl)isoquinoline derivatives VUF5455 and VUF8504. The Ki values of various compounds for [125I]I-AB-MECA binding are listed in Table 1. VUF5455 and VUF8504, compounds with similar allosteric potencies, had the lowest and highest affinities for human A3 adenosine receptors, respectively.
Fig. 5.
Competition of [125I]I-AB-MECA binding to human A3 adenosine receptors expressed in HEK293 cells by VUF5455 andVUF8504. Binding of 0.15 nM [125I]I-AB-MECA to membranes of HEK293 cells expressing human A3 adenosine receptors (25 μg of protein) was carried out in duplicate at 37°C for 1 h in 100 μl 50 mM Tris-HCl buffer, pH 8.0, containing 10 mM MgCl2, 1 mM EDTA, and 0.1 mg/ml CHAPS. The reaction was terminated by rapid filtration through GF/B filters by using a Brandel cell harvester. The figure shows representative experiments with VUF5455 and VUF8504.
Subtype-Selectivity of the Test Compounds as Allosteric Enhancers for A3 Adenosine Receptors
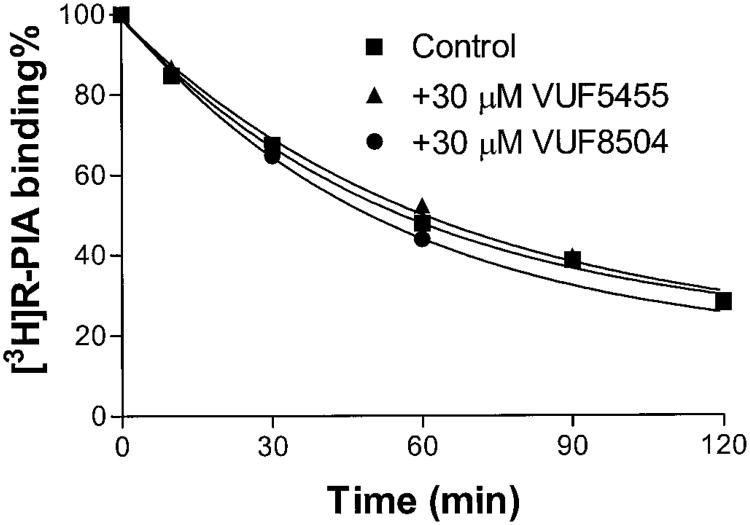
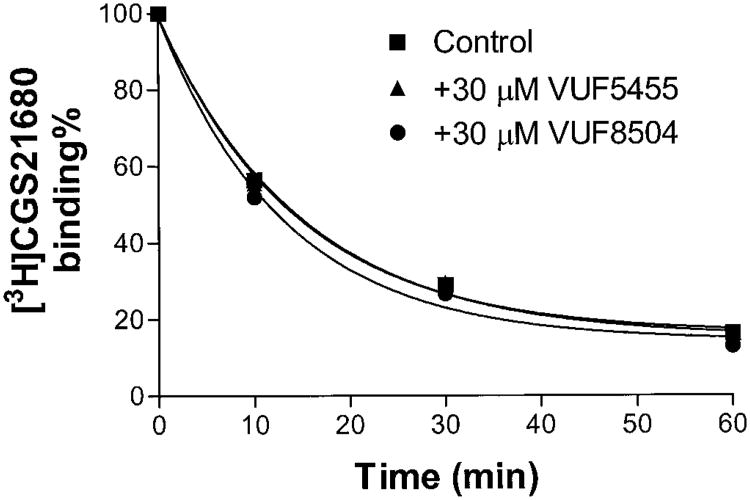
It was demonstrated above that these compounds decreased the dissociation rate of [125I]I-AB-MECA from A3 adenosine receptors. An important issue was whether these compounds were selective for A3 adenosine receptors. To address this issue, we used [3H]R-PIA to investigate whether agonist dissociation from A1 adenosine receptors was altered by these modulators. VUF5455 (30 μM) had no effect on the dissociation rate of [3H]R-PIA from A1 adenosine receptors (Fig. 6). The enhancers also had no effect on the time course for the dissociation of [3H]CGS21680 from A2A adenosine receptors (Fig. 7). These results suggest that the 3-(2-pyridinyl)isoquinoline derivatives are selective allosteric modulators for A3 adenosine receptors.
Fig. 6.
Time course of the dissociation from [3H]R-PIA dissociation from rat A1 adenosine receptors. Binding of 1 nM [3H]R-PIA to A1 adenosine receptors in rat forebrain membranes was carried out at 37°C for 90 min 50 mM Tris-HCl buffer, pH 7.7, in a total assay volume of 400 μl. The dissociation was begun by addition of 10 μM CPA in the absence or presence of the tested compound. Nonspecific binding was determined using 10 μM CPA. Samples were filtered after incubation at 37°C at the time points indicated. The data shown were derived from one experiment performed in duplicate and are typical of three independent experiments giving similar results.
Fig. 7.
Time course of dissociation of [3H]CGS21680 from rat A2A adenosine receptors. Rat striatal membranes (80 μg) were incubated with 15 nM [3H]CGS21680 at 25°C for 90 min in 400 μl of 50 mM Tris-HCl, pH 7.7, containing 10 mM MgCl2. NECA (10 μM) was used to define nonspecific binding. Dissociation was started by the addition of 10 μM NECA in the presence and absence of tested compounds. The data shown were from one experiment performed in duplicate and are representative of three independent experiments giving similar results.
Structure-Activity Relationships for the Enhancement of A3 Adenosine Receptor Binding by 3-(2-Pyridinyl)isoquinoline Derivatives
All of the test compounds that displayed allosteric effects contained a carbonyl group. The replacement of the carbonyl group with an imino group resulted in a loss of the allosteric property of these compounds. The allosteric potencies of the 3-(2-pyridinyl)isoquinoline derivatives VUF5455, VUF8502, VUF8504, and VUF8507 were of the same order of magnitude. By comparison, the order of potencies by which these compounds displaced the binding of [125I]I-AB-MECA to A3 adenosine receptors was: VUF8504 > VUF8502 > CGS15943 > VUF8507 > VUF8505 > VUF8503 > VUF8501 > VUF5455. The compound VUF8504 was about 2 orders of magnitude more potent than VUF5455 with respect to competitive binding.
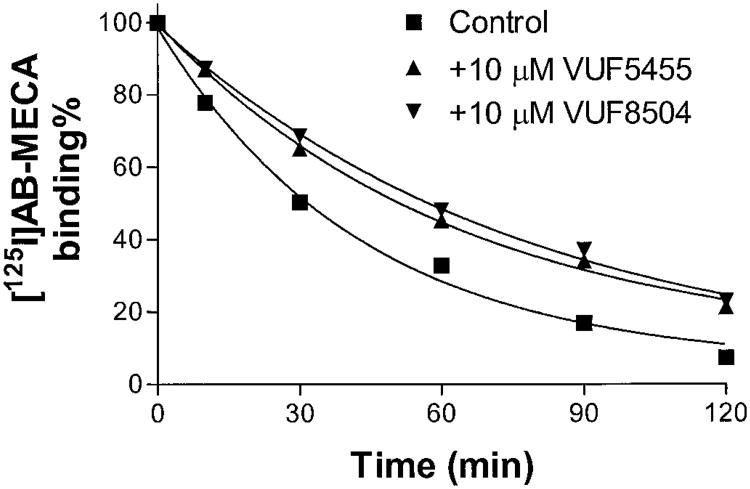
Both the allosteric and competitive potencies of these compounds for rat A3 adenosine receptors were tested. As shown in Fig. 8, the effect of VUF5455 and VUF8504 on the dissociation of [125I]I-AB-MECA from rat A3 adenosine receptors was only slightly decreased or similar to that of human A3 adenosine receptors. In contrast, their competitive binding affinities were approximately 8- and 240-fold less than at human A3 receptors, for VUF5455 (Ki = 12.8 ± 3.1 μM) and VUF8504 (Ki = 4.2 ± 0.8 μM), respectively. This demonstrated that binding enhancement could be separated from competitive antagonism.
Fig. 8.
Time course of the dissociation of [125I]I-AB-MECA dissociation from rat A3 adenosine receptors. [125I]I-AB-MECA (0.15 nM) was preassociated with the rat A3 adenosine receptor membranes (60 μg of protein) at 37°C for 1 h. Nonspecific binding was determined in parallel by addition of 30 μM NECA before the preassociation. After 1-h preassociation, dissociation was initiated by addition of NECA (final concentration, 30 μM) and tested agents. The incubation was terminated by filtration through GF/B filters on a Brandel cell harvester. The data shown were derived from one experiment performed in duplicate and are typical of three independent experiments giving similar results.
Enhancement of the A3 Adenosine Receptor Function by VUF5455
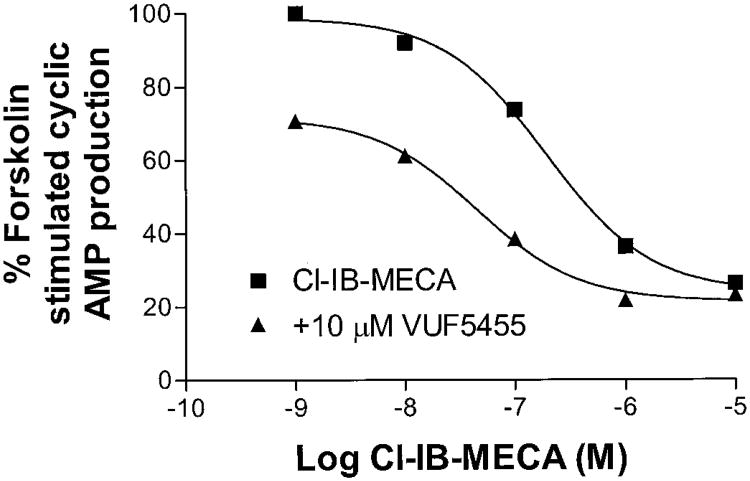
Direct information regarding the allosteric modulation of A3 adenosine receptors was obtained from dissociation kinetic experiments. It was important to correlate data from in vitro assays with those estimated from functional experiments. Hence, we conducted a cAMP assay to observe the effects of the allosteric modulator on human A3 adenosine receptor function. The compound VUF5455 was selected for the cAMP assay, because its competitive binding to A3 adenosine receptors was relatively weak compared with other analogs. VUF5455 significantly enhanced the effect of Cl-IB-MECA on forskolin-induced cAMP formation. The EC50 value of Cl-IB-MECA induced inhibition of forskolin-stimulated cAMP accumulation was 11.7 ± 2.3 nM. In the presence of 1 and 3 μM VUF5455, the EC50 values were 7.4 ± 1.2 and 6.3 ± 0.8 nM, respectively, which were significantly different from that in the absence of VUF5455 (p < 0.05). It should be noted that VUF5455 might exert both allosteric enhancement and antagonistic effects in a functional assay. To avoid the confusing influence by the competitive inhibition, the effect of VUF5455 on the concentration-response curve for Cl-IB-MECA was investigated in the presence of the competitive antagonist MRS1220. Under this experimental condition, 10 μM VUF5455 caused a 3.2-fold shift to the left in the concentration-response relationship for Cl-IB-MECA (Fig. 9). The EC50 values obtained were 232 ± 67.4 and 72.5 ± 18.4 nM (n = 3) in the absence and presence of 10 μM VUF5455, respectively, which were significantly different (p < 0.05).
Fig. 9.
Effect of VUF5455 on the inhibition of forskolin-stimulated cAMP production in HEK293 cells by the A3 receptor agonist Cl-IB-MECA. Forskolin (10 μM) was used to stimulate cAMP levels. MRS1220 (100 nM) was used in the experiment to overwhelm the competitive effect by VUF5455. All experiments were performed in the presence of 10 μM rolipram and 3 U/ml adenosine deaminase. Figure is from a single experiment and is representative of three experiments performed in duplicate.
Discussion
This study provided evidence that several 3-(2-pyridinyl)i-soquinoline derivatives were allosteric enhancers of the A3 adenosine receptor. The allosteric interaction was shown by the slowing of the dissociation of the agonist radioligand [125I]I-AB-MECA from A3 adenosine receptors.
The 3-(2-pyridinyl)isoquinoline derivatives were discovered as potential antagonists for A3 adenosine receptors (van Muijlwijk-Koezen et al., 1998). Previous studies demonstrated that the 3-(2-pyridinyl)isoquinoline derivative VUF8505 was a moderately potent and selective competitive antagonist for A3 adenosine receptors (van Muijlwijk-Koezen et al., 1998). The present study further demonstrated that this compound acts on human A3 adenosine receptors via the competitive binding site, but not an allosteric site.
VUF8504 was approximately 2 orders of magnitude more potent than VUF5455 in displacing [125I]I-AB-MECA binding, whereas the allosteric potencies of the two compounds were similar. A benzamide carbonyl group on the isoquinoline derivatives (Fig. 1A) was required for allosteric enhancement of A3 agonist binding, whereas the corresponding imines (Fig. 1B) did not enhance A3 agonist binding. These results showed that there were distinct structural requirements for allosteric enhancement of A3 adenosine receptor binding, and these requirements were different from those for competitive A3 antagonistic activity. For these two derivatives, species differences in A3 antagonist potency were pronounced, whereas the allosteric effects were comparable in rat and human. Apparently, it was possible to achieve some separation between allosteric and antagonistic activity of these compounds.
The compounds described in this study may serve as the first generation of allosteric enhancers for A3 adenosine receptors. Allosteric enhancers of A3 adenosine receptors may be of potential clinical use; however, application of the modulators as described in this study may have several limitations. An obvious factor that may limit the use of these agents is their antagonistic activities, which will tend to suppress any enhancing actions of these compounds. However, it seems that the structure-activity relationships for allosteric enhancement are separable from those for competitive antagonism, suggesting that it may be possible to discover compounds with improved enhancing activity that lack antagonist activity. Other limitations of these compounds include their low aqueous solubility (around 50 μM) and their modest allosteric potencies. Furthermore, the specificity of these compounds for adenosine receptors compared with other receptors has not been investigated. Hence, the possibility of other interfering activities cannot be excluded.
The identification of allosteric sites on receptors offers new pharmacological approaches to modulate receptor function. Although it is not known whether an allosteric site is a general feature of the GPCR family, increasing numbers of GPCRs are reported to be modulated allosterically by certain compounds. These include: metabotropic glutamate (Litschig et al., 1999), Ca2+ (Conigrave et al., 2000), P2Y (Nepveu et al., 1998; Conigrave et al., 2000), muscarinic (Holzgrabe and Mohr, 1998), adenosine (Kourounakis et al., 2000), adrenergic (Leppik et al., 2000), dopamine (Hoare et al., 2000), 5-hydroxytryptamine (Thomas et al., 1997), tachykinin (Knaus et al., 1991; Croci et al., 1998), angiotensin AT1 (Purdy et al., 1993), and oxytocin receptors (Grazzini et al., 1998). Although no GPCR allosteric enhancer is presently in clinical use, much progress has been made toward the allosteric modulation of this receptor family. Muscarinic receptor researchers currently use the radiolabeled allosteric ligand W84 as a probe to characterize the allosteric site on that receptor (Tränkle et al., 1998). Allosteric enhancers were found with almost absolute selectivity for certain subtypes of muscarinic receptors (Birdsall et al., 2001). This subtype selectivity was considered very difficult to achieve otherwise by targeting the recognition site on the receptor (Tucek and Proska, 1995). The allosteric enhancers of adenosine receptors have a potential advantage as therapeutic agents over adenosine receptor agonists [i.e., they seem less likely to cause desensitization and down-regulation of the receptor (Bhattacharya and Linden, 1996)]. The concept that an allosteric recognition site might serve as an alternate means to effect receptor activation was proposed and demonstrated for several G protein-coupled receptors, including muscarinic (Jakubik et al., 1996), 5-hydroxytryptamine7 (Thomas et al., 1997), P2Y (Conigrave et al., 2000), and Ca2+ receptors (Conigrave et al., 2000; Kobilka, 2000).
In the absence of mutation data for the allosteric effect, it is not yet possible to locate the allosteric site or even to speculate about the distance between and uniqueness of allosteric and orthosteric ligand binding sites on the A3 receptor. Nevertheless, there may be some structural interdependency between the two sites, which is indicated also by the fact that the same chemical class of ligands may bind at both. For the A1 receptor, a single mutation of T277A, had effects on both (i.e., the affinity of agonists greatly diminished and the effect of PD81723 was much less pronounced) (Kourounakis et al., 2001). The nature and location of the allosteric sites on muscarinic receptors have not been characterized in detail; however, mutation studies showed that residues found important for the binding of gallamine were mainly located at the extracellular face of the receptor. Some of these residues might be part of one of the allosteric sites (Birdsall et al., 2001). The fact that many allosteric modulators for muscarinic receptors are polycationic molecules, together with other evidence, suggests that the allosteric site for some modulators may be close to the orthosteric site. The allosteric site of muscarinic receptors is thought to be near the exofacial surface of the receptor (Christopoulos et al., 1998).
In summary, allosteric modulators for A3 adenosine receptors were identified and characterized. All of the compounds that showed allosteric enhancement also showed competitive binding properties. However, structure-activity relationships suggested the allosteric effects were separable from competitive antagonism. Hence, it may be possible to find allosteric enhancers that lack antagonistic properties by modifying the structures of these compounds or by screening compounds of other chemical entities.
Acknowledgments
We thank Miriam Dissen, Jacobien von Frijtag Drabbe Künzel, and Neli Melman for their assistance with the ligand binding experiments.
Z.-G.G. received financial support from Gilead Sciences (Foster City, CA).
Abbreviations
- GPCR
G protein-coupled receptor
- I-AB-MECA
N6-(4-amino-3-iodobenzyl)-5′ -N-methylcarboxamidoadenosine
- R-PIA
N6-[(R)-phenylisopropyl]adenosine
- CGS21680
2-[p-(2-carboxyethyl)phenyl-ethylamino]-5′-N-ethylcarboxamidoadenosine
- BCA
bicinchoninic acid
- CGS15943
5-amino-9-chloro-2-(2-furyl)-1,2,4-triazolo[1,5-c]quinazoline
- CPA
N6-cyclopentyladenosine
- NECA
5′-N-ethylcarboxamidoadenosine
- HEK
human embryonic kidney
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfornate
- VUF5455
4-methoxy-N-[7-methyl-3-(2-pyridinyl)-1-isoquinolinyl]benzamide
- VUF8502
4-methyl-N-[3-(2-pyridinyl)-1-isoquinolinyl]benzamide
- VUF8504
4-methoxy-N-[3-(2-pyridinyl)-1-isoquinolinyl]benzamide
- VUF8507
N-[3-(2-pyridinyl)-1-isoquinolinyl]benzamide
- VUF8501
N-[3-(2-pyridinyl)-1-isoquinolinyl]benzenecarboximidamide
- VUF8503
4-methyl-N-[3-(2-pyridinyl)-1-isoquinolinyl]benzenecarboximidamide
- VUF8505
4-methoxy-N-[3-(2-pyridinyl)-1-isoquinolinyl]benzenecarboximidamide
- PD81723
2-amino-4,5-dimethyl-3-thienyl-[3-(trifluoromethyl)phenyl]methanone
- PSB-11
8-ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H-imidazo[2.1-i]purin-5-one
References
- Bhattacharya S, Linden J. Effects of long-term treatment with the allosteric enhancer, PD81,723, on Chinese hamster ovary cells expressing recombinant human A1 adenosine receptors. Mol Pharmacol. 1996;50:104–111. [PubMed] [Google Scholar]
- Birdsall NJM, Cohen F, Lazareno S, Matsui H. Allosteric regulation of G-protein-linked receptors. Biochem Soc Trans. 1995;23:108–111. doi: 10.1042/bst0230108. [DOI] [PubMed] [Google Scholar]
- Birdsall NJ, Lazareno S, Popham A, Saldanha J. Multiple allosteric sites on muscarinic receptors. Life Sci. 2001;68:2517–2524. doi: 10.1016/s0024-3205(01)01047-5. [DOI] [PubMed] [Google Scholar]
- Bruns RF, Fergus JH. Allosteric enhancement of adenosine A1 receptor binding and function by 2-amino-3-benzoylthiophenes. Mol Pharmacol. 1990;38:939–949. [PubMed] [Google Scholar]
- Christopoulos A, Lanzafame A, Mitchelson F. Allosteric interactions at muscarinic cholinoceptors. Clin Exp Pharmacol Physiol. 1998;25:185–194. doi: 10.1111/j.1440-1681.1998.t01-4-.x. [DOI] [PubMed] [Google Scholar]
- Conigrave AD, Quinn SJ, Brown EM. l-Amino acid sensing by the extracellular Ca2+-sensing receptor. Proc Natl Acad Sci USA. 2000;97:4814–4819. doi: 10.1073/pnas.97.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci T, Aureggi G, Manara L, Emonds-Alt X, LeFur G, Maffrand JP, Mukenge S, Ferla G. In vitro characterization of tachykinin NK2-receptors modulating motor responses of human colonic muscle strips. Br J Pharmacol. 1998;124:1321–1327. doi: 10.1038/sj.bjp.0701960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredholm BB, Arslan G, Halldner L, Kull B, Schulte G, Wasserman W. Structure and function of adenosine receptors and their genes. Naunyn Schmiede-berg's Arch Pharmacol. 2000;362:364–374. doi: 10.1007/s002100000313. [DOI] [PubMed] [Google Scholar]
- Gao ZG, IJzerman AP. Allosteric modulation of A2A adenosine receptors by amiloride analogues and sodium ions. Biochem Pharmacol. 2000;60:669–676. doi: 10.1016/s0006-2952(00)00360-9. [DOI] [PubMed] [Google Scholar]
- Gao ZG, Jiang Q, Jacobson KA, IJzerman AP. Site-directed mutagenesis studies of human A2A adenosine receptors. Involvement of Glu13 and His278 in ligand binding and sodium modulation. Biochem Pharmacol. 2000;60:661–668. doi: 10.1016/s0006-2952(00)00357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazzini E, Guillon G, Mouillac B, Zingg HH. Inhibition of oxytocin receptor function by direct binding of progesterone. Nature (Lond) 1998;392:509–512. doi: 10.1038/33176. [DOI] [PubMed] [Google Scholar]
- Hannon JP, Pfannkuche HJ, Fozard JR. A role for mast cells in adenosine A3 receptor-mediated hypotension in the rat. Br J Pharmacol. 1995;115:945–952. doi: 10.1111/j.1476-5381.1995.tb15902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare SRJ, Coldwell MC, Armastrong D, Strange PG. Regulation of human D1, D2(long), D2(short), D3 and D4 dopamine receptors by amiloride and amiloride analogues. Br J Pharmacol. 2000;130:1045–1059. doi: 10.1038/sj.bjp.0703370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzgrabe U, Mohr K. Allosteric modulators of ligand binding to muscarinic acetylcholine receptors. Drug Disc Today. 1998;3:214–222. [Google Scholar]
- Jakubik J, Bacakova L, Lisa V, El-Fakahany EE, Tucek S. Activation of muscarinic acetylcholine receptors via their allosteric binding sites. Proc Natl Acad Sci USA. 1996;93:8705–8709. doi: 10.1073/pnas.93.16.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SM, Quinn RJ. Adenosine receptors as potential therapeutic targets. Drug Disc Today. 1999;4:542–551. doi: 10.1016/s1359-6446(99)01421-x. [DOI] [PubMed] [Google Scholar]
- Knaus GA, Knaus HG, Saria A. Allosteric interaction of heparin with NK-1 receptors in rat striatal membranes. Ann NY Acad Sci. 1991;632:422–425. doi: 10.1111/j.1749-6632.1991.tb33143.x. [DOI] [PubMed] [Google Scholar]
- Kobilka B. Allosteric activation of CaR by l-amino acids. Proc Natl Acad Sci USA. 2000;97:4419–4420. doi: 10.1073/pnas.97.9.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourounakis AP, van der Klein PAM, IJzerman AP. Elucidation of structure-activity relationships 2-amino-3-benzoylthiophenes: Study of their allosteric enhancing vs. antagonistic activity on adenosine A1 receptors. Drug Dev Res. 2000;49:227–237. [Google Scholar]
- Kourounakis A, Visser C, de Groote M, IJzerman AP. Differential effects of the allosteric enhancer (2-amino-4,5-dimethyl-trienyl)[3-trifluoromethyl) phenyl]methanone (PD81,723) on agonist and antagonist binding and function at the human wild-type and a mutant (T277A) adenosine A1 receptor. Biochem Pharmacol. 2001;61:137–144. doi: 10.1016/s0006-2952(00)00536-0. [DOI] [PubMed] [Google Scholar]
- Lee HT, Emala CW. Protective effects of renal ischemic preconditioning and adenosine pretreatment: role of A1 and A3 receptors. Am J Physiol Renal Physiol. 2000;278:F380–387. doi: 10.1152/ajprenal.2000.278.3.F380. [DOI] [PubMed] [Google Scholar]
- Leppik RA, Mynett A, Lazareno S, Birdsall NJM. Allosteric interactions between the antagonist prazosin and amiloride analogs at the human α1A-adrenergic receptor. Mol Pharmacol. 2000;57:436–445. doi: 10.1124/mol.57.3.436. [DOI] [PubMed] [Google Scholar]
- Linden J. Allosteric enhancement of adenosine receptors. In: Jacobson KA, Jarvis MF, editors. Purinergic Approaches in Experimental Therapeutics. Wiley-Liss Inc.; New York: 1997. pp. 85–97. [Google Scholar]
- Litschig S, Gasparini F, Rueegg D, Stoehr N, Flor PJ, Vranesic I, Prézeau L, Pin JP, Thomsen C, Kuhn R. CPCCOEt, a noncompetitive metabotropic glutamate receptor antagonist, inhibits receptor signaling without affecting glutamate binding. Mol Pharmacol. 1999;55:453–461. [PubMed] [Google Scholar]
- MacKenzie WM, Hoskin DW, Blay J. Adenosine inhibits the adhesion of anti-CD3-activated killer lymphocytes to adenocarcinoma cells through an A3 receptor. Cancer Res. 1994;54:3521–3526. [PubMed] [Google Scholar]
- Müller CE. A3 adenosine receptor antagonists. Mini-Reviews in Med Chem. 2001 doi: 10.2174/1389557510101040417. In press. [DOI] [PubMed] [Google Scholar]
- Nepveu F, Souchard JP, Rolland Y, Dorey G, Spedding M. 2–2′-Pyridylisatogen, a selective allosteric modulator of P2 receptors, is a spin trapping agent. Biochem Biophys Res Commun. 1998;242:272–276. doi: 10.1006/bbrc.1997.7949. [DOI] [PubMed] [Google Scholar]
- Olah ME, Gallo-Rodriguez C, Jacobson KA, Stiles GL. 125I-4-aminobenzyl-5′-N-methylcarboxamidoadenosine, a high affinity radioligand for rat A3 adenosine receptor. Mol Pharmacol. 1994;45:978–982. [PMC free article] [PubMed] [Google Scholar]
- Purdy RE, Prins BA, Weber MA, Bakhtiarian A, Smith JR, Kim MK, Nguyen THT, Wiler EWJ. Possible novel action of ouabain: allosteric modulation of vascular serotonergic (5-HT2) and angiotensinergic (AT1) receptor. J Pharmacol Exp Ther. 1993;267:228–237. [PubMed] [Google Scholar]
- Sajjadi FG, Takabayashi K, Foster AC, Domingo RC, Firestein GS. Inhibition of TNF-α expression by adenosine: role of A3 adenosine receptors. J Immunol. 1996;156:3435–3442. [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Strickler J, Jacobson KA, Liang BT. Direct preconditioning of cultured chick ventricular myocytes. Novel functions of cardiac adenosine A2a and A3 receptors. J Clin Invest. 1996;98:1773–1779. doi: 10.1172/JCI118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA, Carson MJ, Neal MJ, Sutcliffe JG. Unique allosteric regulation of 5-hydroxytryptamine receptor-mediated signal transduction by oleamide. Proc Natl Acad Sci USA. 1997;94:14115–14119. doi: 10.1073/pnas.94.25.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tränkle C, Mies-Klomfass E, Cid MHB, Holzgrabe U, Mohr K. Identification of a [3H]ligand for the common allosteric site of muscarinic acetylcholine M2 receptors. Mol Pharmacol. 1998;54:139–145. doi: 10.1124/mol.54.1.139. [DOI] [PubMed] [Google Scholar]
- Tucek S, Proska J. Allosteric modulation of muscarinic acetylcholine receptors. Trends Pharmacol Sci. 1995;16:205–212. doi: 10.1016/s0165-6147(00)89023-9. [DOI] [PubMed] [Google Scholar]
- van Muijlwijk-Koezen JE, Timmerman H, Link R, van der Goot H, IJzerman AP. A novel class of adenosine A3 receptor ligands. 1. 3-(2-Pyridinyl)isoquinoline derivatives. J Med Chem. 1998;41:3987–3993. doi: 10.1021/jm980036q. [DOI] [PubMed] [Google Scholar]
- Von Lubitz DK, Lin RC, Popik P, Carter MF, Jacobson KA. Adenosine A3 receptor stimulation and cerebral ischemia. Eur J Pharmacol. 1994;263:59–67. doi: 10.1016/0014-2999(94)90523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QY, Li C, Olah ME, Johnson RA, Stiles GL, Civelli O. Molecular cloning and characterization of an adenosine receptor: the A3 adenosine receptor. Proc Natl Acad Sci USA. 1992;89:7432–7436. doi: 10.1073/pnas.89.16.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]