Abstract
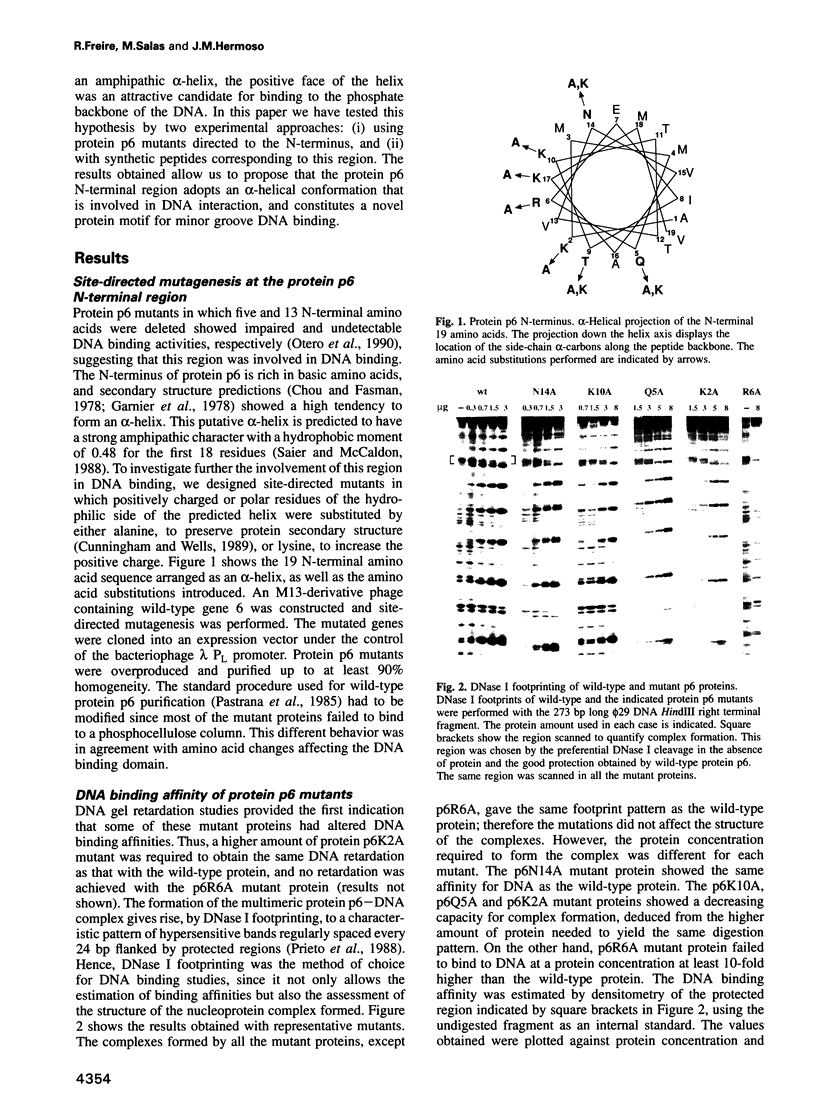
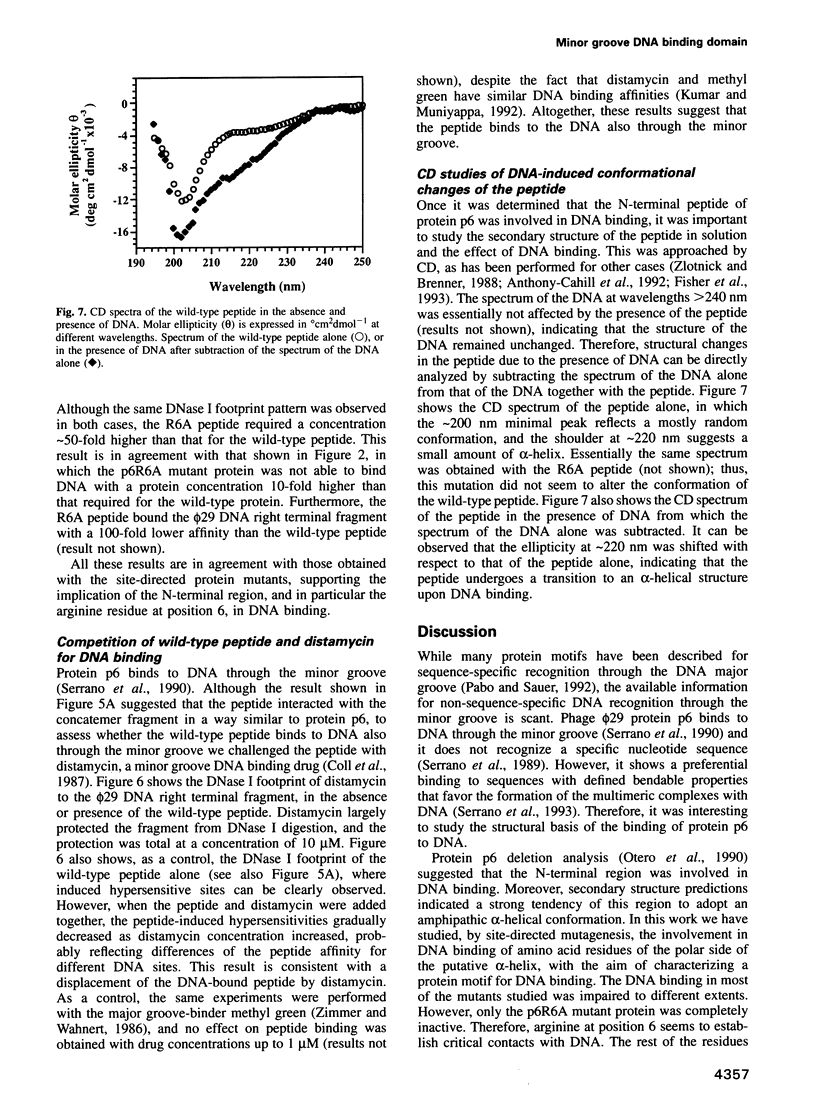
Protein p6 of the Bacillus subtilis phage phi 29 binds with low sequence specificity to DNA through the minor groove, forming a multimeric nucleoprotein complex that activates the initiation of phi 29 DNA replication. Deletion analysis suggested that the N-terminal part of protein p6, predicted to form an amphipathic alpha-helix, is involved in DNA binding. We have constructed site-directed mutants at the polar side of the putative alpha-helix. DNA binding and activation of initiation of phi 29 DNA replication were impaired in most of the mutant proteins obtained. A 19 amino acid peptide comprising the N-terminus of protein p6 interacted with a DNA fragment containing high-affinity signals for protein p6 binding with approximately 50-fold higher affinity than the peptide corresponding to an inactive mutant. Both wild-type peptide and protein p6 recognized the same sequences in this DNA fragment. This result, together with distamycin competition experiments, suggested that the wild-type peptide also binds to DNA through the minor groove. In addition, CD spectra of the wild-type peptide showed an increase in the alpha-helical content when bound to DNA. All these results indicate that an alpha-helical structure located in the N-terminal region of protein p6 is involved in DNA binding through the minor groove.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony-Cahill S. J., Benfield P. A., Fairman R., Wasserman Z. R., Brenner S. L., Stafford W. F., 3rd, Altenbach C., Hubbell W. L., DeGrado W. F. Molecular characterization of helix-loop-helix peptides. Science. 1992 Feb 21;255(5047):979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- Balzer D., Ziegelin G., Pansegrau W., Kruft V., Lanka E. KorB protein of promiscuous plasmid RP4 recognizes inverted sequence repetitions in regions essential for conjugative plasmid transfer. Nucleic Acids Res. 1992 Apr 25;20(8):1851–1858. doi: 10.1093/nar/20.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Salas M. Transition from initiation to elongation in protein-primed phi 29 DNA replication: salt-dependent stimulation by the viral protein p6. J Virol. 1988 Nov;62(11):4167–4172. doi: 10.1128/jvi.62.11.4167-4172.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Gutiérrez J., Lázaro J. M., Bernad A., Salas M. Replication of phage phi 29 DNA in vitro: role of the viral protein p6 in initiation and elongation. Nucleic Acids Res. 1986 Jun 25;14(12):4923–4937. doi: 10.1093/nar/14.12.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M. A., Lázaro J. M., Blanco L., Salas M. Phi 29 DNA polymerase active site. The conserved amino acid motif "Kx3NSxYG" is involved in template-primer binding and dNTP selection. J Biol Chem. 1993 Aug 5;268(22):16763–16770. [PubMed] [Google Scholar]
- Carrascosa J. L., Camacho A., Moreno F., Jiménez F., Mellado R. P., Viñuela E., Salas M. Bacillus subtilis phage phi29. Characterization of gene products and functions. Eur J Biochem. 1976 Jul 1;66(2):229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989 Jun 2;244(4908):1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- Fisher D. E., Parent L. A., Sharp P. A. High affinity DNA-binding Myc analogs: recognition by an alpha helix. Cell. 1993 Feb 12;72(3):467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Giese K., Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994 Mar;10(3):94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
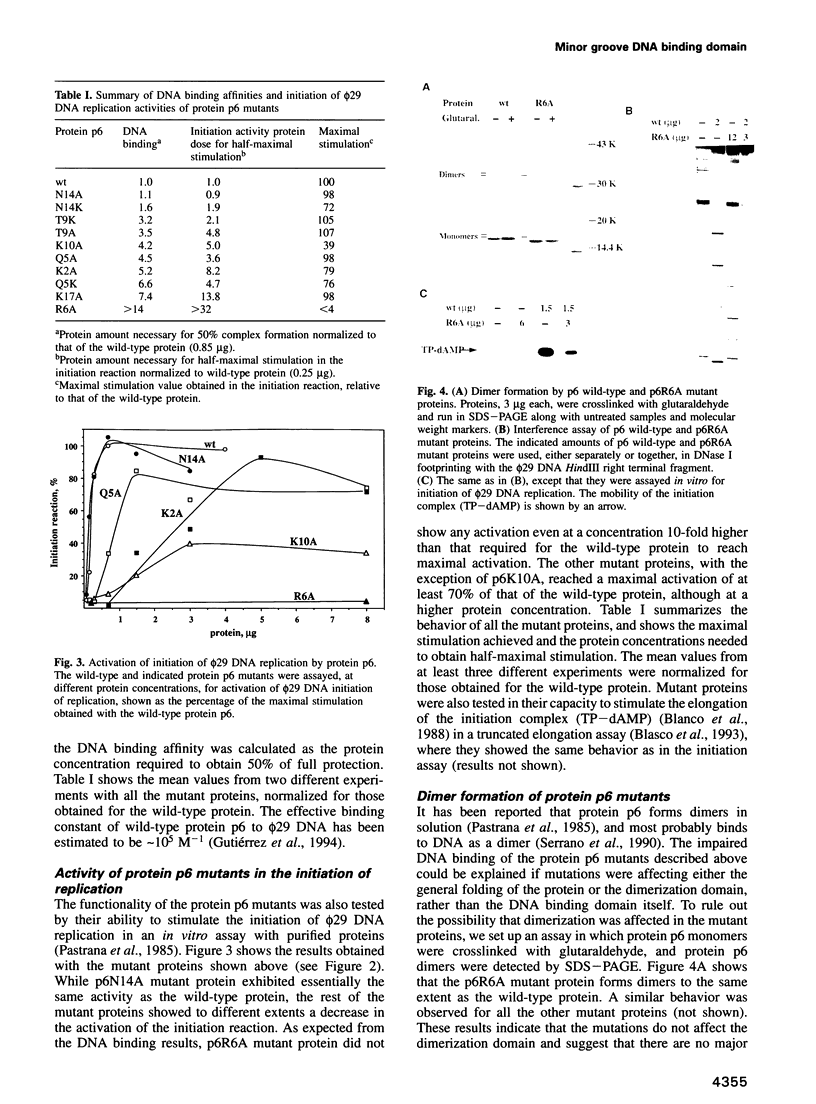
- Gutiérrez C., Freire R., Salas M., Hermoso J. M. Assembly of phage phi 29 genome with viral protein p6 into a compact complex. EMBO J. 1994 Jan 1;13(1):269–276. doi: 10.1002/j.1460-2075.1994.tb06257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- King C. Y., Weiss M. A. The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: a topological mechanism of sequence specificity. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11990–11994. doi: 10.1073/pnas.90.24.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K. A., Mahalakshmi S., Muniyappa K. DNA-induced conformational changes in RecA protein. Evidence for structural heterogeneity among nucleoprotein filaments and implications for homologous pairing. J Biol Chem. 1993 Dec 15;268(35):26162–26170. [PubMed] [Google Scholar]
- Kumar K. A., Muniyappa K. Use of structure-directed DNA ligands to probe the binding of recA protein to narrow and wide grooves of DNA and on its ability to promote homologous pairing. J Biol Chem. 1992 Dec 5;267(34):24824–24832. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Mellado R. P., Peñalva M. A., Inciarte M. R., Salas M. The protein covalently linked to the 5' termini of the DNA of Bacillus subtilis phage phi 29 is involved in the initiation of DNA replication. Virology. 1980 Jul 15;104(1):84–96. doi: 10.1016/0042-6822(80)90367-0. [DOI] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Otero M. J., Lázaro J. M., Salas M. Deletions at the N terminus of bacteriophage phi 29 protein p6: DNA binding and activity in phi 29 DNA replication. Gene. 1990 Oct 30;95(1):25–30. doi: 10.1016/0378-1119(90)90409-k. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Pastrana R., Lázaro J. M., Blanco L., García J. A., Méndez E., Salas M. Overproduction and purification of protein P6 of Bacillus subtilis phage phi 29: role in the initiation of DNA replication. Nucleic Acids Res. 1985 May 10;13(9):3083–3100. doi: 10.1093/nar/13.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva M. A., Salas M. Initiation of phage phi 29 DNA replication in vitro: formation of a covalent complex between the terminal protein, p3, and 5'-dAMP. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto I., Serrano M., Lázaro J. M., Salas M., Hermoso J. M. Interaction of the bacteriophage phi 29 protein p6 with double-stranded DNA. Proc Natl Acad Sci U S A. 1988 Jan;85(2):314–318. doi: 10.1073/pnas.85.2.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon B. J., Winters R. S., Gordon D., Winter E. A peptide motif that recognizes A.T tracts in DNA. Proc Natl Acad Sci U S A. 1993 Dec 1;90(23):11327–11331. doi: 10.1073/pnas.90.23.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Nissen M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990 May 25;265(15):8573–8582. [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, McCaldon P. Statistical and functional analyses of viral and cellular proteins with N-terminal amphipathic alpha-helices with large hydrophobic moments: importance to macromolecular recognition and organelle targeting. J Bacteriol. 1988 May;170(5):2296–2300. doi: 10.1128/jb.170.5.2296-2300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Gutiérrez C., Salas M., Hermoso J. M. Superhelical path of the DNA in the nucleoprotein complex that activates the initiation of phage phi 29 DNA replication. J Mol Biol. 1993 Mar 5;230(1):248–259. doi: 10.1006/jmbi.1993.1140. [DOI] [PubMed] [Google Scholar]
- Serrano M., Gutiérrez J., Prieto I., Hermoso J. M., Salas M. Signals at the bacteriophage phi 29 DNA replication origins required for protein p6 binding and activity. EMBO J. 1989 Jun;8(6):1879–1885. doi: 10.1002/j.1460-2075.1989.tb03584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Salas M., Hermoso J. M. A novel nucleoprotein complex at a replication origin. Science. 1990 May 25;248(4958):1012–1016. doi: 10.1126/science.2111580. [DOI] [PubMed] [Google Scholar]
- Story R. M., Weber I. T., Steitz T. A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992 Jan 23;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Gerstein M., Johnson T. An NMR study on the DNA-binding SPKK motif and a model for its interaction with DNA. Protein Eng. 1993 Aug;6(6):565–574. doi: 10.1093/protein/6.6.565. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell W. G., Satchwell S. C., Travers A. A. A decapeptide motif for binding to the minor groove of DNA. A proposal. FEBS Lett. 1988 May 23;232(2):263–268. doi: 10.1016/0014-5793(88)80750-6. [DOI] [PubMed] [Google Scholar]
- Warrant R. W., Kim S. H. alpha-Helix-double helix interaction shown in the structure of a protamine-transfer RNA complex and a nucleoprotamine model. Nature. 1978 Jan 12;271(5641):130–135. doi: 10.1038/271130a0. [DOI] [PubMed] [Google Scholar]
- White S. W., Appelt K., Wilson K. S., Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5(4):281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]
- Zlotnick A., Brenner S. L. An alpha-helical peptide model for electrostatic interactions of proteins with DNA. The N terminus of RecA. J Mol Biol. 1989 Oct 5;209(3):447–457. doi: 10.1016/0022-2836(89)90009-0. [DOI] [PubMed] [Google Scholar]