Fig. 6.
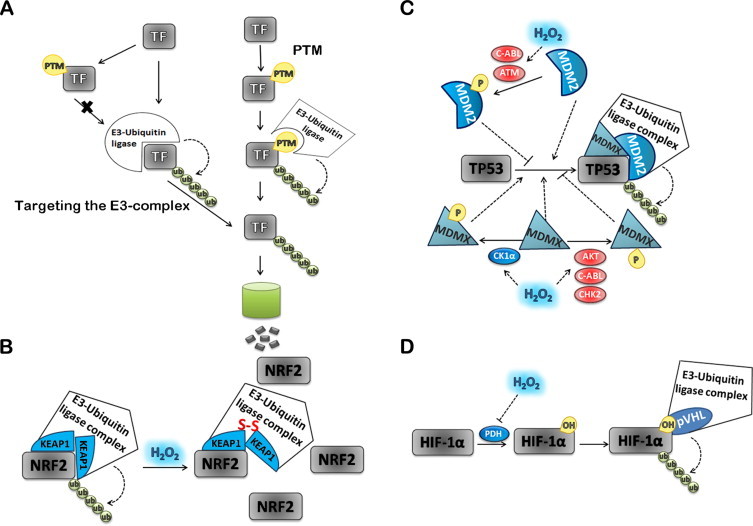
Regulation of TFs activity by degradation modulated by H2O2. TF degradation via poliubiquitin(Ub)-proteasome pathway is an important regulatory mechanism in different signaling pathways (A). Proteasome, a multicatalytic protease oligomeric complex, degrades proteins that have been poly-ubiquitinated. Ubiquitin (Ub)-protein ligase (E3) enzymes transfer the activated Ub from an Ub-conjugating protein E2, first to a lysine residue of the substrate protein and the next Ub to lysine residues present in the last added ubiquitin, originating an Ub chain. NRF2 ubiquitination and degradation involves KEAP1 as the substrate adaptor subunit in the E3 holoenzyme (B). In the presence of H2O2 critical cysteine residues in KEAP1 are oxidized, changing KEAP1 conformation. This conformational change affects the interaction between NRF2 and KEAP1 inhibiting NRF2 ubiquitination and degradation. In normal cells TP53 is maintained at low levels by interaction with the negative regulator MDM2 a p53 highly specific ubiquitin ligase. H2O2 mainly regulates MDM2 activity by activating the ATM and c-ABL kinases involved in MDM2 phosphorylation. MDM2 phosphorylation inhibits its ubiquitination activity and stabilizes TP53 (C). The MDM2 ubiquitin ligase substrate preference for TP53 is enhanced by MDMX. Phosphorylation of MDMX in different residues by c-ABL, AKT and CHK2 kinases inhibit TP53 degradation, while those catalyzed by the CK1α kinase stimulates TP53 degradation. All the referred kinases activities are regulated by H2O2. The association between the TF and the ubiquitination machinery can also be modulated by PTMs of the transcription factor (D). HIF-1α is tagged with ubiquitin for degradation after being hydroxylated by PHD in the presence of O2 (D). H2O2 inhibits PHD that leads to HIF-1α stabilization. Factors colored blue are inhibitors of TF-dependent gene expression; factors colored red are activators of TF-dependent gene expression. Dashed lines indicate activation/inhibition.
