Figure 1.
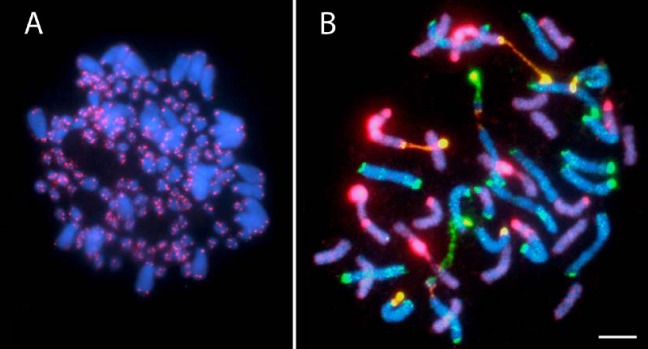
ISH to root tip metaphase spreads (A) FISH to pentaploid Agave fourcroydes (2n = 5x = 150) probed with telomere repeat sequences (TTAGGG)n (biotin labelled probe Cy3 signal, pink), chromosomes counterstained with DAPI (blue). (B) GISH to a partial metaphase spread of a Nicotiana sylvestris (2n = 2x = 24) x N. tomentosiformis (2n = 2x = 24) allotetraploid, which has a chromosome complement of 2n = 4x = 48 (see Kostoff hybrid [2]). The spread is probed with total genomic DNA from N. sylvestris (digoxigenin-labelled probe, FITC signal, green) and from N. tomentosiformis (biotin-labelled probe, Cy3 signal, pink), chromosomes are counterstained with DAPI (blue). Depending on the mixture and intensity of the fluorophores other colours are generated. Chromosomes of N. sylvestris origin are shown as cyan and green depending on the strength of the FITC/DAPI signal, while those of N. tomentosiformis origin are violet and magenta depending on the strength of the Cy3/DAPI signal. Areas of yellow chromatin represent 35S ribosomal DNA (rDNA) loci that are hybridised with genomic DNA from both parents. Scale bar: 10 µm.
