Abstract
A role for vitamin A in host defense against Mycobacterium tuberculosis has been suggested through epidemiological and in vitro studies; however, the mechanism is unclear. Here, we demonstrate that vitamin A-triggered antimicrobial activity against M. tuberculosis requires expression of Niemann-Pick disease type C2 (NPC2). Comparison of monocytes stimulated with all-trans retinoic acid (ATRA) or 1,25-dihydroxyvitamin D3 (1,25D3), the biologically active forms of vitamin A and vitamin D, respectively, indicates that ATRA and 1,25D3 induce mechanistically distinct antimicrobial activities. Stimulation of primary human monocytes with ATRA did not result in expression of the antimicrobial peptide cathelicidin, which is required for 1,25D3 antimicrobial activity. In contrast, ATRA triggers a reduction in the total cellular cholesterol concentration, whereas 1,25D3 did not. Blocking ATRA-induced cellular cholesterol reduction inhibits antimicrobial activity as well. Bioinformatic analysis of ATRA and 1,25D3 induced gene profiles suggests Niemann-Pick disease type C2 (NPC2) is a key gene in ATRA-induced cholesterol regulation. Knockdown experiments demonstrate that ATRA-mediated decrease of total cellular cholesterol content and increase in lysosomal acidification are both dependent upon expression of NPC2. Expression of NPC2 was lower in caseous tuberculosis granulomas and M. tuberculosis-infected monocytes compared to normal lung and uninfected cells, respectively. Loss of NPC2 expression ablated ATRA-induced antimicrobial activity. Taken together, these results suggest that the vitamin A-mediated antimicrobial mechanism against M. tuberculosis requires NPC2-dependent expression and function, indicating a key role for cellular cholesterol regulation in the innate immune response.
INTRODUCTION
One key function of the innate immune system is the rapid recognition and destruction of invading pathogens via the activation of antimicrobial pathways. In the case of Mycobacterium tuberculosis, the causative agent of tuberculosis, micronutrients have proven to be critical as part of a successful antimicrobial response. Epidemiological evidence demonstrates an association between vitamin A and tuberculosis: serum vitamin A levels are significantly higher in healthy household contacts compared to tuberculosis patients (1,2). In the laboratory, the biologically active form of vitamin A, all-trans retinoic acid (ATRA), has been shown to inhibit growth of virulent M. tuberculosis in macrophages (3,4). However, the molecular mechanisms and cellular processes induced by ATRA that lead to this antimicrobial activity are unclear.
Vitamin D and vitamin A share similar molecular and biochemical characteristics: both are fat-soluble secosteroids that are recognized by and that effect changes in cells by binding to the vitamin D receptor (VDR) and the retinoic acid receptor (RAR), respectively (5). RAR and VDR are members of the nuclear hormone receptor family and heterodimerize with the retinoid X receptor (RXR) (5). In relation to tuberculosis, deficient serum vitamin D levels are associated with tuberculosis (6,7) and treatment of M. tuberculosis infected cells in vitro with the active 1,25-dihydroxyvitamin D3 (1,25D3) form of vitamin D triggers antimicrobial activity (8,9), which is comparable to the epidemiological and biochemical properties of vitamin A. Based on these similarities, we compared the 1,25D3-triggered antimicrobial response, which is dependent on production of the antimicrobial peptide cathelicidin (10,11), to the ATRA-triggered response in order to elucidate the vitamin A-mediated antimicrobial mechanism.
MATERIALS AND METHODS
Statistical analysis
Comparisons between two different conditions were analyzed using Student's t-test. All experiments with three or more measurements were analyzed using One Way ANOVA or Kruskal-Wallis One Way ANOVA on Ranks as appropriate with Student-Newman-Keuls Method for pairwise analyses. Error bars represent the standard error of the mean.
Reagents
All-trans-retinoic acid (ATRA) was purchased (Sigma-Aldrich, St. Louis, MO), dissolved in DMSO and stored at −80°C in small aliquots protected from light. 1,25-dihydroxyvitamin D3 (1,25D3) was purchased (Enzo Life Sciences, Farmingdale, NY, USA), dissolved in ethanol and stored at −80°C in small aliquots protected from light. Both the ATRA and 1,25D3 were utilized at 10−8M. Lysosensor Green DND 189 (Life Technologies) was used at 1:2000 dilution (0.5 nM) as recommended by the manufacturer. Oregon-Green-488-dextran MW10,000 and Alexa-Fluor-647-dextran MW10,000 were purchased (Life Technologies) and used at 250 μg/mL and at 30 μg/mL, respectively. Nelfinavir and Ritonavir were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID.
Cell Culture
This study was conducted according to the principles expressed in the Declaration of Helsinki, and was approved by the Institutional Review Board of the University of California at Los Angeles. All donors provided written informed consent for the collection of peripheral blood and subsequent analysis. We obtained whole blood from healthy donors through the UCLA CFAR Virology Core with informed consent. Mononuclear cells (PBMCs) were isolated from peripheral blood of healthy donors using Ficoll-Paque as previously described (12,13). Monocytes were purified using two methods: plastic adherence and negative selection. For plastic adherence, PBMCs were cultured for two hours in 1640 RPMI medium (Life Technologies) supplemented with 1% fetal calf serum (Omega Scientific, Tarzana, CA, USA). The cultures were vigorously washed, and the remaining adherent cells were cultured in RPMI with 10% fetal calf serum. For negatively selected monocytes, we utilized EasySep Human Monocyte Enrichment Kit (StemCell Technologies, Vancouver, BC, Canada) according to the manufacturer's recommended protocol. Monocyte-derived macrophages (MDMs) were produced as previously described with M-CSF (14).
M. tuberculosis
M. tuberculosis H37Ra and H37Rv were plated on 7H11 agar plates from frozen stocks. All experiments involving H37Rv were conducted at Biosafety Level 3. Following three to four weeks of incubation at 37°C in a water jacketed incubator with 5% CO2, the solid colonies were scraped off the agar plate and placed in 1xPBS. The bacterial suspension was gently separated with a sonicating water bath (Branton 2510) for 30 seconds and then centrifuged at 735g for four minutes to create a single cell suspension. To enumerate the bacteria, the supernatant was separated from the pellet and the absorbance at 600nm was measured using spectrophotometry. Normal monocytes and MDMs were infected at an MOI of one and transfected monocytes at an MOI of 0.5 overnight, and then the cells are vigorously washed three times with fresh RPMI media to remove extracellular bacteria.
Antimicrobial assay
In order to assess M. tuberculosis viability from infected monocytes, we utilized the real time PCR based method as previously described (15,16), which compares 16S RNA levels to a genomic DNA (IS6110) levels as an indicator of bacterial viability. Monocytes were purified and infected with M. tuberculosis and stimulated as indicated for three days. For H37Ra-infected monocytes, the cells are harvested and divided following the incubation. Half of the cells were lysed by boiling at 100°C for 5 minutes then snap frozen at −80°C. Total RNA was isolated from the remaining half using TRIzol (Life Technologies) via phenol-chloroform extraction, followed by RNA cleanup and on-column DNAse digestion using RNeasy Miniprep Kit (Qiagen, Valencia, CA). cDNA was synthesized from the total RNA using the iScript cDNA Synthesis kit (BioRad, Hercules, CA) according to the manufacturer's recommended protocol. The bacterial 16S rRNA, and genomic element DNA levels were then assessed from the cDNA and cellular lysate, respectively, using real time PCR using iQ SYBR Green (BioRad). Comparison of the bacterial DNA to the mammalian genomic 36B4 levels was used to monitor infectivity between all the conditions in the assay as well as PCR quality. The relative 16S values were calculated using the ΔΔCT analysis, with the IS6110 value serving as the “housekeeping gene”. The IS6110 genomic element and 16S primer sequences are as follows: 16S Forward 5'-GGT GCG AGC GTT GTC CGG AA-3', 16S Reverse 5'- CGC CCG CAC GCT CAC AGT TA-3', IS6110 Forward 5'-GGA AGC TCC TAT GAC AAT GCA CTA G-3', and IS6110 Reverse 5'-TCT TGT ATA GGC CGT TGA TCG TCT-3'. For H37Rv infected cells, the DNA was isolated from the interphase and phenol-chloroform phase using the back extraction protocol as described by the manufacturer.
Quantitative real-time RT-PCR for mRNA
Total RNA was extracted from cells using Trizol (Life Technologies) and mRNA was reverse transcribed using iScript (Bio-Rad). Gene expression of CAMP, CYP27A1, NPC2 and IL6 were analyzed by quantitative real-time RT-PCR (qPCR) as described above with 36B4 as the housekeeping gene. The primers are as follows: CAMP Forward 5'-GGA CCC AGA CAC GCC AAA-3', CAMP Reverse 5'-GCA CAC TGT CTC CTT CAC TGT GA-3', CYP27A1 Forward 5'-GCT ATG CCC TGC AAC TGC ACC A-3', CYP27A Reverse 5'-TCC TTC CGT GGT GAA CGG CCC ATA G-3', NPC2 Forward 5'-TAT CCC TCT ATA AAA CTG GTG GTG-3', NPC2 Reverse 5'-CCA GAT GCA CCG AAC TCA AT-3', IL6 Forward 5'-GCC CAC CGG GAA CGA AAG AGA-3', IL6 Reverse 5'-GAC CGA AGG CGC TTG TGG AGA AG-3'.
Cellular Cholesterol Measurement
Monocytes were cultured and stimulated as indicated for 18 hours, then collected and enumerated. The lipid fraction was extracted using 3:2 hexane:isopropyl alcohol at room temperature for 30 min. Following 10 min centrifugation the supernatant was collected and dried in glass test tubes using nitrogen gas. Cholesterol levels were then assessed using the Amplex Red Cholesterol Assay Kit (Life Technologies) using the recommended protocol and expressed as total cholesterol per cell.
Microarray analysis
Total RNA was isolated from monocytes treated as indicated in the Results section. The total RNA samples were processed and analyzed by the UCLA Clinical Microarray Core Facility using the Affymetrix U133 Genechip. Cluster diagrams were generated using the Cluster and TreeView software programs from the Eisen Lab at http://rana.lbl.gov/ (17). Biological functions, cholesterol related functions were identified using Ingenuity Pathways Analysis (http://www.ingenuity.com/). For the caseous tuberculosis granuloma and WGCNA validation microarray analysis, data files were obtained from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/, accession numbers GSE20050, GSE23073, GSE13762, and GSE28995) (18). NPC2 and IL6 levels were normalized to G3PDH. Since there are multiple G3PDH probes represented on the microarray, the NPC2 and IL6 probe values were normalized to every G3PDH probe and averaged.
Weighted gene co-expression network analysis (WGCNA)
We performed WGCNA as previously described, via the R package “WGCNA.” (19). A signed weighted correlation network was constructed using the blockwiseModules() command with a soft thresholding power β = 9. Using an adjacency matrix calculated from pairwise correlations between all pairs of genes across all samples raised to the power β, the topological overlap was calculated to measure of network interconnectedness. Module eigengenes were calculated for each resulting module, and were correlated to ATRA or 1,25D3 treatment using a binary vector representation of treatment status.
WGCNA Module preservation
WGCNA can assess whether individual modules are preserved between two data sets (19). To validate our microarray results, we examined the preservation our ATRA- and 1,25D3- specific modules against published and publically available microarray data (20–22). The R function `modulePreservation' in the WGCNA R package was applied to our data and the published data, and the Zsummary value was calculated. Zsummary scores >10 are interpreted as “strongly preserved”, Zsummary scores between 2 and 10 are interpreted as “weak to moderately preserved”, and Zsummary scores <2 are “not preserved”. Significance of the Zsummary scores are calculated by permutation analysis (23).
Identification of hub genes
Genes with highest module membership values, or module eigengene based connectivity (kME), are referred to as intramodular “hub” genes, which are genes that have the highest number of connections within the module. kME was calculated for each gene in each module using the signedKME() command. For each module, genes were sorted by kME and VisANT was used to visualize the gene connections among the top 50hub genes as ranked by kME (24).
Monocyte viability
Two methods were used to determine monocyte viability: Trypan Blue exclusion and TUNEL assay. Following infection for 16 hours with H37Ra, monocytes were harvested and viability was assessed. For Trypan Blue exclusion the harvested cell were incubated with a final concentration of 0.04% Trypan Blue (Biorad), and enumerated for the number of blue labeled cells compared to total cells using an automated cell counter (Biorad TC10). The data is represented as a percentage of viable cells. For TUNEL assay, the APO-BrdU TUNEL Assay Kit (Life Technologies) was utilized with the manufacturer's recommended protocol.
Transfection of monocytes
Purified monocytes were transfected with siRNA oligos using the Lonza Nucleofector 4D system with the Human Monocyte Nucleofector kit (Lonza, Allendale, NJ) according to the manufacturer's recommended protocol. To knockdown expression of NPC2, the pre-designed and validated ON-TARGETplus siRNA oligo pool (siNPC2) was utilized in conjunction with the ON-TARGETplus Non Targeting Pool (siCTRL) control. The siRNA oligos were purchased and stored as recommended by the manufacturer (Thermo Fisher, Rockford, IL, USA).
Lysosomal acidification
Lysosomal acidification was measured using two dyes: LysoSensor (Life Technologies) and Oregon-Green (Life Technologies) as previously described (25). Monocytes were purified and transfected as described above with siCTRL or siNPC2, then stimulated with ATRA for 18 hours. LysoSensor (1:2000) was added to each well and rocked gently for five seconds to mix then incubated for 30 minutes. After incubation, the cells were fixed in 1% PFA and fluorescence was acquired using flow cytometry. The mean fluorescent intensity was determined on the monocytes in the samples, which were the only cellular population in the samples given that they were magnetically separated as indicated in the “Transfection of monocytes” section above. For Oregon-Green labeling, the transfected and stimulated monocytes were harvested, washed and incubated with Oregon-Green-488 at 250 μ/mL and Alexa-Fluor-647 (Life Technologies) at 30 μ/mL for 30 minutes. The cells were washed and analyzed by flow cytometry. Both Oregon-Green-488 and Alexa-Fluor-647 were conjugated to 10,000MW dextran for targeting into the lysosome. Data shown is the ratio of the fluorescence detected for Oregon-Green-488 to Alexa-Fluor-647.
RESULTS
ATRA- and 1,25D3-induced monocyte function
Primary human monocytes were infected overnight with M. tuberculosis H37Ra, an avirulent strain used to model mycobacterial infection (10,15), and then treated with carrier control, ATRA or 1,25D3 for three days. Following treatment, the ratio of bacterial 16S RNA to the IS6110 genomic repeat element was determined as an indicator of bacterial viability as we previously described (15). Both ATRA and 1,25D3 treatment resulted in a decrease of bacterial viability as compared to carrier control (representative figure, Figure 1A). On average, M. tuberculosis viability, measured as log10(16S/IS6110) in monocytes treated with ATRA or 1,25D3, was significantly reduced (Figure 1B) and of comparable magnitude at these concentrations (10−8M) to previously published studies using macrophages and the colony forming unit (CFU) assay (3,8–10).
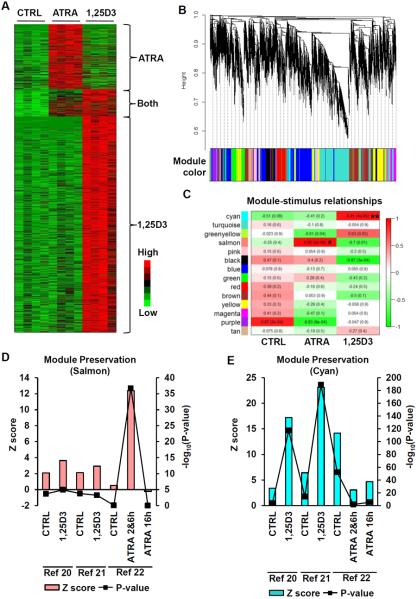
Figure 1.
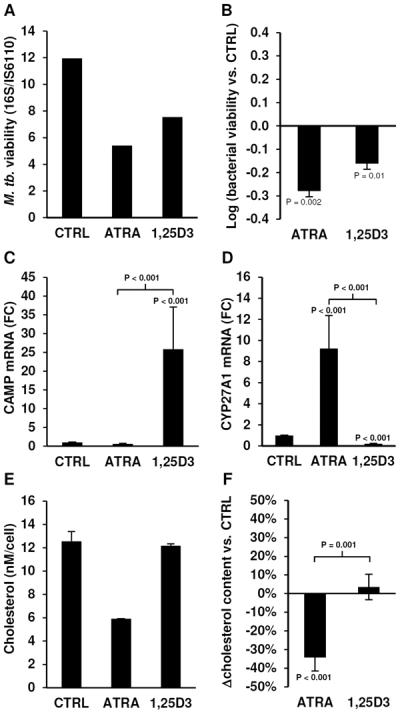
ATRA- and 1,25D3-induced cellular responses. Bacterial viability of M. tuberculosis H37Ra infected primary monocytes treated with carrier control (CTRL), ATRA or 1,25D3 for three days displayed as (A) a representative experiment or (B) log10(mean bacterial viability vs. CTRL ± SEM), n = 4. mRNA expression levels assessed by qPCR of (C) cathelicidin (CAMP) and (D) CYP27A1 in primary human monocytes stimulated with CTRL, ATRA or 1,25D3 for 18 hours, displayed as mean fold change vs. CTRL ± SEM, n = 7. Cellular cholesterol levels per cell of monocytes treated with CTRL, ATRA or 1,25D3 for 18 hours shown as (E) a representative experiment or (F) mean percent change vs. CTRL ± SEM, n = 5.
Given that the 1,25D3-mediated antimicrobial activity is dependent on expression of the antimicrobial peptide cathelicidin (10,11), we determined if ATRA could induce cathelicidin (CAMP) mRNA in primary human monocytes. Although 1,25D3 was able to significantly induce expression of CAMP mRNA, ATRA did not (Figure 1C). In contrast, CYP27A1 mRNA, a known ATRA response gene (26), was induced by ATRA but not 1,25D3 (Figure 1D), indicating that the cells were responding to ATRA. Since cholesterol is a key factor in the interactions between innate immune cells and M. tuberculosis (27–29), and ATRA is known to induce cholesterol efflux (26,30,31), we hypothesized that the ability of ATRA to regulate cellular cholesterol content is linked to antimicrobial activity. The cellular cholesterol concentration (total cholesterol/cell) in monocytes treated with carrier control, ATRA or 1,25D3 was measured (representative figure, Figure 1E). ATRA treatment resulted in a significant reduction (−34% ± 7% vs. control, P < 0.001) of total cellular cholesterol content, whereas 1,25D3 had no effect (Figure 1F). These results suggest that ATRA and 1,25D3 induce distinct intracellular pathways, and likely utilize different antimicrobial mechanisms.
Role of cholesterol regulation in ATRA-induced antimicrobial activity
To determine if ATRA-induced reduction in cellular cholesterol plays a role in ATRA-mediated antimicrobial activity, we utilized Nelfinavir (Nel) and Ritonavir (Rit), two compounds previously described to inhibit cholesterol efflux from human macrophages and macrophage-derived foam cells (32,33). Monocytes were infected with M. tuberculosis H37Ra for 18 hours, washed to remove extracellular bacterium, preincubated for 10 minutes with Nel at 10μ-M and Rit at 30μ-M, which are concentrations previously described to inhibit cholesterol efflux (32,33), and then stimulated with ATRA for three days. ATRA induced a decrease in cellular cholesterol compared to carrier control, which was significantly inhibited by the presence of Nel or Rit (Figure 2A). There were no significant effects of Nel or Rit on baseline total cellular cholesterol levels (Supplemental Figure 1A). Importantly, ATRA-induced antimicrobial activity was significantly inhibited in M. tuberculosis H37Ra-infected monocytes by Nel (from −1.3 to −0.05 ± 0.17, P = 0.0002 vs. ATRA only) and Rit (from −1.3 to −0.13 ± 0.22, P = 0.002 vs. ATRA only) (Figure 2B).
Figure 2.
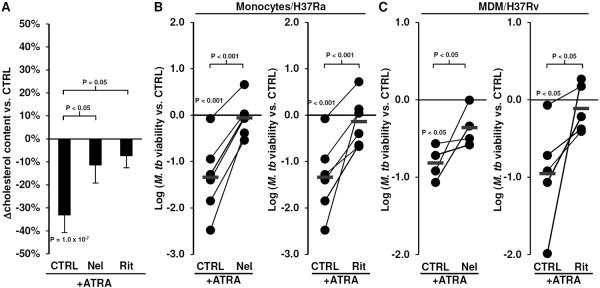
Effects of of Nelfinavir (Nel) and Ritonavir (Rit) on ATRA-induced responses. (A) Monocytes treated with Nel and Rit for 20 minutes then stimulated with ATRA for 18 hours. The total cellular cholesterol level per cell was determined using Amplex Red. Data shown is the mean percent change vs. CTRL treated cells ± SEM, n = 5. Bacterial viability of (B) M. tuberculosis H37Ra in monocytes or (C) M. tuberculosis H37Rv in monocyte-derived macrophages (MDMs) treated with ATRA with or without Nel or Rit for three days. Data shown is log10(bacterial viability vs. CTRL) for each individual donor monocytes/MDMs tested. Red line indicates the mean.
Since macrophages are the natural cellular host type for M. tuberculosis infection, we utilized monocyte-derived macrophages (MDMs), a cellular model for antimicrobial activity against M. tuberculosis in macrophages (14). MDMs were infected with the virulent M. tuberculosis H37Rv strain at a MOI of one for 18 hours, washed to remove extracellular bacteria, preincubated for 10 minutes with Nel and Rit, and then stimulated with ATRA for three days. Pretreatment M. tuberculosis H37Rv-infected MDMs with Nel or Rit inhibited ATRA-induced antimicrobial activity (Nel; from −0.9 to −0.05 ± 0.16, P = 0.05 vs. ATRA only) (Rit; from −0.85 to −0.28 ± 0.15, P = 0.05 vs. ATRA only) (Figure 2C). These data are suggestive that regulation of cellular cholesterol may play an important role in ATRA-induced antimicrobial activity in both monocytes and macrophages.
ATRA- vs. 1,25D3-induced gene expression profiles
To identify specific genes driving the ATRA-induced cholesterol regulation, we compared the monocyte gene expression profiles induced by ATRA (10−8M) or 1,25D3 (10−8M) treatment for 18 hours from four independent donors using microarrays. Analysis of genes significantly upregulated (1.2 fold vs. control, P < 0.05) by either ATRA or 1,25D3 revealed three gene groups: i) induced by ATRA only, ii) induced by 1,25D3 only, and iii) induced by both (Figure 3A). A total of 868 genes were represented in the ATRA only group, 2591 genes in the 1,25D3 only group, and 205 genes induced by both. As expected, CYP27A1 was significantly upregulated (2.6 ± 0.3 fold vs. control, P < 0.001) and present in the ATRA only group (Supplemental Figure 1B).
Figure 3.
ATRA- and 1,25D3-induced gene expression profiles. (A) Hierarchical clustering of genes induced (1.2 fold over CTRL, P < 0.05) in primary human monocytes by ATRA or 1,25D3 after 18 hours. (B) Identification of co-expression modules by WGCNA. The dendrogram was obtained by average linkage hierarchical clustering of ATRA and 1,25D3 gene expression profiles. Height on the dendrogram represents kME, with more connected genes located towards the bottom of the tree. The corresponding module colors for each gene are indicated in the color bar. (C) Correlation of module eigengenes vs. stimulus displayed in a heatmap. The r-value (P value) are indicated in the map for each module. * and ** denote modules most highly and significantly correlated with ATRA or 1,25D3 treatment, respectively. Preservation (Zscore) of the (D) Salmon and (E) Cyan modules in published microarray studies on ATRA or 1,25D3 stimulated immune cells, P values indicated by line graph overlay.
To confirm the differential gene expression signatures induced by ATRA or 1,25D3, we applied Weighted Gene Co-expression Network Analysis (WGCNA) to the microarray data (34). WGCNA identified transcripts that organize into distinct modules of co-expressed genes (Figure 3B). In particular, the “salmon” module eigengene was significantly correlated with ATRA stimulation (r = 0.95, P = 2 × 10−6), whereas the “cyan” module eigengene was correlated with 1,25D3 stimulation (r = 0.91, P = 4 × 10−5) (Figure 3C). To validate these ATRA- and 1,25D3-induced gene expression profiles, we determined the preservation of the cyan and salmon modules in published and publically available microarray studies that examined: CD14+ monocytes differentiating into dendritic cells treated with 1,25D3 (10−8M) for 18 hours (20), human primary monocytes treated with 1,25D3 (10−8M) for 12 hours (21) and the THP-1 monocytic cell line treated with ATRA (2 × 10−8M) for two, six and 16 hours (22). These studies were chosen for the comparison to our current study because they i) were conducted on myeloid immune cells, ii) used similar doses of ATRA and 1,25D3, and iii) utilized similar incubation times. Due to the low number of THP-1 samples at two (n = 2) and six (n = 2) hours for the ATRA study, the data from these two time points were combined in order to provide the necessary resolution for the module preservation test. The cyan module demonstrated a strong preservation in the 1,25D3 studies (Figure 3D) whereas the salmon module was only preserved in the ATRA study (Figure 3E). These results suggest that the gene expression profiles we obtained monocytes represent distinct core gene signatures induced by ATRA and 1,25D3.
Identification of candidate genes
To determine the relationship between the gene expression profiles and cellular function, the upregulated genes were analyzed by Ingenuity Pathway Analysis (IPA), a knowledge-guided bioinformatics tool, to identify biological functions enriched by ATRA or 1,25D3 (analysis scheme displayed in Supplemental Figure 2). Based on our hypothesis that control of cellular lipids is a key element of the ATRA-induced antimicrobial response, the categories of “lipid metabolism”, “molecular transport” and “small molecule biochemistry” (the second, third and fourth ranked functions, respectively) were further examined (Figure 4A). The same three categories (“lipid metabolism”, “molecular transport” and “small molecule biochemistry”) were the 27th, 23rd and 2nd ranked 1,25D3-induced categories, respectively (Figure 4B). Comparing the ATRA- and 1,25D3-induced genes in the `small molecular biochemistry' category reveals 14 genes in common, which represents 6.2% of the total genes in the category induced by either ATRA or 1,25D3 (Supplemental Figure 2). Taken together, these analyses suggest that ATRA induces a lipid metabolism and intracellular molecular transport gene profile that is not present in 1,25D3-stimulated monocytes.
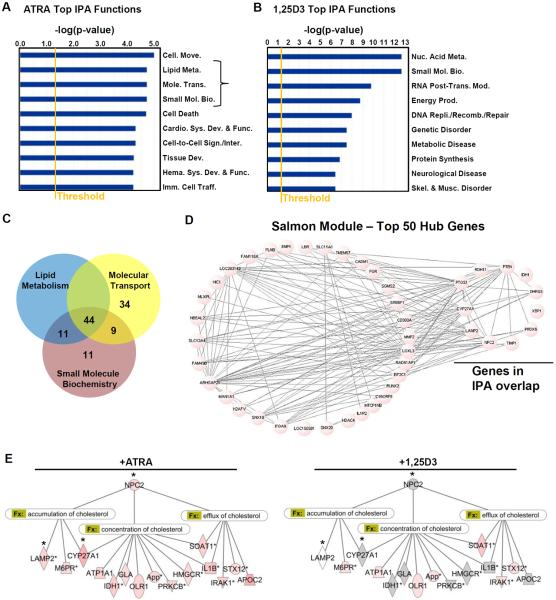
Figure 4.
Regulation of cellular cholesterol by ATRA. Ingenuity Pathways Analysis (IPA) of the top biological functions induced by (A) ATRA or (B) 1,25D3. Intensity of red indicates level of significance (p < 0.05, Fishers Exact Test), grey indicates not significant for ATRA- or 1,25D3-induced expression. (C) Venn diagram of the ATRA-induced genes in the “lipid metabolism”, “molecular transport” and “small molecule biochemistry” categories. (D) Network connection between the top 50 ranked hub genes identified by WGCNA. The overlap between the 44 common genes and the hub genes are indicated. (E) Annotation of the 44 common genes with IPA functional categories (Fx) related to cholesterol. * indicates WGCNA identified hub genes.
The comparison of the ATRA-induced genes in the “lipid metabolism”, “molecular transport” and “small molecule biochemistry” categories revealed a high degree of similarity between the categories, with 44 genes in common (Figure 4C). A total of 11 genes out of the 44 common genes were also identified as hub genes by WGCNA (Figure 4D). Of the 44 common genes, 16 genes were annotated by IPA with functions related to regulation of cellular cholesterol (functions: “accumulation of cholesterol”, “concentration of cholesterol”, and “efflux of cholesterol”), three of which (NPC2, CYP27A1 and LAMP2) were also hub genes. Only one gene, Niemann-Pick disease type C2 (NPC2), was annotated with all three cholesterol-related functions, and was induced by ATRA (Figure 4E) but not by 1,25D3 (Figure 4F). The kME value of NPC2 was 0.98 which was the highest ranked hub gene identified by WGCNA in the salmon module. These data suggest that NPC2 may play a central role in the ATRA-induced gene signature that mediates the intracellular regulation of cholesterol content.
Induction of NPC2 by ATRA
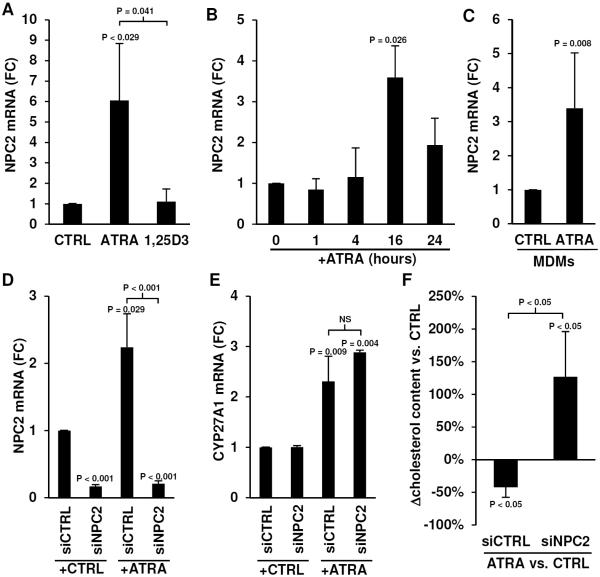
Monocytes were stimulated with ATRA or 1,25D3 and NPC2 mRNA levels were measured by qPCR. Confirming the microarray analysis, the qPCR results demonstrated specific induction of NPC2 mRNA in ATRA stimulated monocytes compared to 1,25D3 stimulation (Figure 5A). To better characterize induction of NPC2, monocytes were stimulated with ATRA at 10−8M for one, four, 16 and 24 hours. Total RNA was harvested and NPC2 mRNA levels were measured by qPCR. NPC2 was significantly induced (3.6 fold vs. control, P = 0.026) at the 16 hour time point in monocytes (Figure 5B). MDMs stimulated with ATRA also showed a significant induction (3.4 fold vs. control, P = 0.008) of NPC2 mRNA at 16 hours (Figure 5C). A retinoic acid receptor response element (RAR-RE) is present 1312 base pairs upstream of the NPC2 mRNA start site (Supplemental Data Figure 3A). These results demonstrate that NPC2, which has a RAR-RE, can be induced by ATRA stimulation of monocytes.
Figure 5.
Regulation of NPC2 by ATRA. mRNA levels of NPC2 assessed by qPCR in (A) primary human monocytes stimulated with carrier control (CTRL) or ATRA or 1,25D3 for 18 hours (mean fold change (FC) vs. CTRL ± SEM, n = 4), (B) monocytes stimulated with ATRA over a time course (mean FC vs. CTRL ± SEM, n = 4, per time point), and (C) human MDMs stimulated with ATRA for 18 hours (mean FC vs. CTRL ± SEM, n = 5). mRNA levels of (D) NPC2 and (E) CYP27A1 in monocytes transfected with a non-targeting siRNA oligo (siCTRL) or a NPC2 specific siRNA oligo (siNPC2), then stimulated with CTRL or ATRA. Data shown is mean FC vs. CTRL ± SEM, n = 3–4. (F) Percent change of total cholesterol per cell of monocytes transfected with siCTRL or siNPC2 then stimulated with ATRA for 18 hours. Data shown is mean percent change vs. CTRL ± SEM, n = 6.
Role of NPC2 in ATRA-induced regulation of cellular cholesterol content
Mutations in NPC1 or NPC2 are associated with Niemann-Pick disease, a lysosomal storage disorder that is characterized by abnormally high cholesterol accumulation in cells. NPC1 and NPC2 have common (35) and non-redundant functions related to lysosomal lipid transport (36,37). Stimulation of monocytes with ATRA resulted in expression of NPC2 but not NPC1 detected by qPCR correlating with the microarray results, suggesting a specific role for NPC2 as a lipid transporter in ATRA-stimulated cells (Supplemental Figure 3B). To ascertain the role of NPC2 in ATRA-mediated regulation of cellular cholesterol concentration, we transfected monocytes with a siRNA oligo specific for NPC2 (siNPC2) or a control non-targeting siRNA oligo (siCTRL), then stimulated the cells with carrier control or ATRA for 18 hours. siNPC2 transfection resulted in a significant decrease of NPC2 mRNA levels in both resting and ATRA-stimulated monocytes (Figure 5D). Neither siNPC2 nor siCTRL had an effect on the CYP27A1 mRNA levels of resting and ATRA-stimulated monocytes (Figure 5E), indicating that the siNPC2 knockdown was specific. Correlating with expression of NPC2, siCTRL-transfected monocytes stimulated with ATRA demonstrated a reduction (−42% ± 15% vs. control treated, P < 0.05) in total cellular cholesterol content (Figure 5F). In marked contrast, ATRA stimulation of siNPC2-transfected monocytes resulted in an increase (126% ± 69% vs. control treated, P < 0.05) in total cellular cholesterol content (Figure 4F). There was no significant difference in baseline cellular cholesterol concentration between untreated siCTRL and siNPC2-transfected cells (Supplemental Figure 3C).
Role of NPC2 in lysosomal acidification
Blocking cholesterol egress from lysosomes prevents acidification (38), which is a key process in antimicrobial activity against M. tuberculosis (39). We sought to address the role of NPC2 in ATRA-induced lysosomal acidification using two dyes: Lysosensor, which accumulates and increases fluorescence intensity in acidic organelles, and Oregon-Green-488, a pH sensitive fluorescent dye (25). siCTRL and siNPC2 transfected monocytes were treated with ATRA for 18 hours, and labeled using Lysosensor. Stimulation with ATRA resulted in increased Lysosensor labeling in siCTRL but not siNPC2 transfected monocytes (Figure 6A). The change in mean fluorescence intensity (MFI) in siCTRL cells treated with ATRA (11.1%, P = 0.012) was significantly higher (P = 0.009) compared to siNPC2 cells (1.2%) treated with ATRA (Figure 6B).
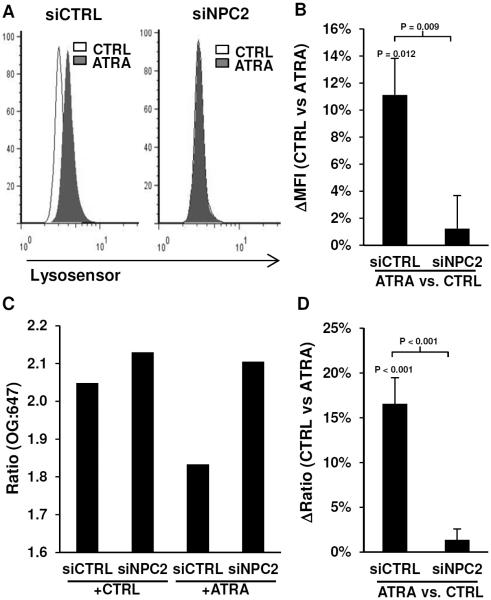
Figure 6.
Lysosomal acidification. Monocytes were transfected with siCTRL or siNPC2 then stimulated with CTRL or ATRA for 18 hours, and lysosomal acidification was measured using Lysosenor or Oregon-Green. Lysosensor data is shown as (A) representative histograms of the flow cytometry data and (B) average change (ATRA vs. CTRL) of the mean fluorescence intensity (ΔMFI) ± SEM, n = 4. Detection of lysosomal acidification by Oregon-Green is calculated as the fluorescence ratio of Oregon-Green-488-dextran (OG) to Alexa-647-dextran (647). The Oregon-Green data is shown as (C) a representative experiment, and (D) average change (ATRA vs. CTRL) of the OG:647 ratio ± SEM, n = 4.
Oregeon-Green-488 is pH sensitive and will exhibit a decrease in fluorescence when exposed to acidic environments, whereas Alexa-647 is pH resistant and the fluorescence will remain constant. Therefore, a decrease in the Oregon-Green-488 to Alexa-647 ratio will indicate an increase in acidification. siCTRL- and siNPC2-transfected monocytes were treated with ATRA for 18 hours, then co-labeled with Oregon-Green-488 and Alexa-647 both conjugated to dextran for lysosomal targeting. Stimulation with ATRA resulted in a decreased Oregon-Green:Alexa ratio (OG:A647) in siCTRL but not siNPC2-transfected monocytes (Figure 6C). The decrease OG:A647 in siCTRL cells treated with ATRA (16.6%, P < 0.001) was significantly higher (P < 0.001) compared to siNPC2 cells (1.4%) treated with ATRA (Figure 6D). These data suggest ATRA induces lysosomal acidification that is dependent upon the expression of NPC2.
Role of NPC2 during M. tuberculosis infection
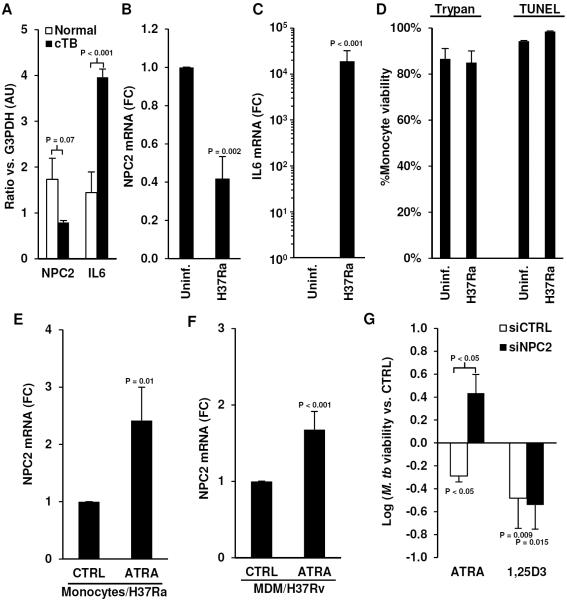
To determine whether M. tuberculosis infection regulates NPC2 expression in situ we analyzed a previously published gene microarray experiment, which compared uninvolved lung tissue to caseous tuberculosis granulomas (40). The relative expression of NPC2 and IL6 was compared to G3PDH in the same samples to account for differences between the sample types. Based on this analysis, NPC2 levels trended lower (0.46 fold, P = 0.07), whereas IL6 levels, which is inducible by M. tuberculosis (41), were significantly higher (2.7 fold, P = 0.0004) in caseous tuberculosis granulomas compared to uninvolved lung tissue (Figure 7A). Since lung biopsies contain multiple cell types, including non-immune cells, the effects of M. tuberculosis infection on the NPC2 signal may be confounded. Therefore, we addressed the effects of M. tuberculosis infection on NPC2 expression directly in monocytes. In monocytes infected with M. tuberculosis H37Ra, we observed a significant decrease (0.4 ± 0.1 fold vs. uninfected, P = 0.002) in NPC2 mRNA after 18 hours (Figure 7B) and an increase in IL6 mRNA (Figure 7C) correlating with the microarray data. There was no change in monocyte viability following infection as determined by Trypan Blue exclusion and TUNEL assay (Figure 7D).
Figure 7.
Role of NPC2 in host defense against M. tuberculosis infection. Expression of (A) NPC2 and IL6 mRNA in caseous tuberculosis granuloma (cTB) vs. normal lung tissue normalized to G3PDH levels by microarray analysis. Expression of (B) NPC2 and (C) IL6 mRNA in M. tuberculosis H37Ra infected monocytes treated with CTRL or ATRA for three days. Data shown is mean FC vs. CTRL ± SEM, n = 5 (D) Monocyte viability following infection assessed with Trypan Blue exclusion and TUNEL assay, data shown is average %viability ± SEM, n = 4. Expression of NPC2 mRNA in (E) M. tuberculosis H37Ra-infected-monocytes (n = 5) or (F) M. tuberculosis-H37Rv-infected MDMs (n = 13) treated with CTRL or ATRA for three days. Data shown is mean FC vs. CTRL ± SEM. (G) Bacterial viability of M. tuberculosis H37Ra in monocytes transfected with siCTRL or siNPC2 then stimulated with ATRA or 1,25D3 for three days. Data shown is log10(mean bacterial viability vs. siCTRL ± SEM), n = 3–4.
Treatment of infected monocytes with ATRA for three days, which parallels the antimicrobial assay time course, resulted in a significant increase of NPC2 mRNA levels in monocytes (Figure 7E) and MDMs (Figure 7F). Knockdown of NPC2 mRNA ablated the ATRA-induced antimicrobial activity (0.29 ± 0.22 vs. siNPC2 control stimulated), whereas transfection of siCTRL had no effect (−0.30 ± 0.08 vs. siCTRL control stimulated, P < 0.05) (Figure 7G). In contrast, knockdown of NPC2 did not affect the ability of 1,25D3 to induce antimicrobial activity (Figure 7G), which is expected based on the fact that NPC2 was not induced by 1,25D3 and that 1,25D3 utilizes a cathelicidin dependent pathway (10). Comparison of siCTRL vs. siNPC2-transfected monocytes treated with vehicle control showed no significant difference in bacterial viability during the course of the antimicrobial activity experiment (Supplemental Figure 3D). Taken together, these findings indicate an important role for NPC2 in the ATRA-triggered antimicrobial response against M. tuberculosis infection.
DISCUSSION
Although vitamin A has been associated with host protection against M. tuberculosis both in vivo (1,2) and in vitro (3,4), the precise vitamin A-induced antimicrobial mechanism remained unclear. Here, we sought to explore the mechanism(s) driving the vitamin A-triggered antimicrobial response by comparing vitamin A (ATRA) and vitamin D (1,25D3) induced cellular and genomic responses, given that both are known to induce antimicrobial activity in M. tuberculosis-infected monocytes and macrophages (3,8,9). Previously, we demonstrated that 1,25D3-induced antimicrobial activity was dependent on expression of the antimicrobial peptide cathelicidin (10,11); however, our current study found that ATRA did not induce cathelicidin expression. In contrast, ATRA, but not 1,25D3 stimulation, resulted in reduction of cellular cholesterol content. Blocking cholesterol egress inhibited ATRA-mediated antimicrobial in monocytes infected with M. tuberculosis H37Ra as well as MDMs infected with M. tuberculosis H37Rv. Bioinformatic analysis combining WGCNA and IPA revealed NPC2, a lysosomal to endoplasmic reticulum lipid transporter, as a potential mediator of ATRA-induced regulation of cellular cholesterol content. NPC2 was required for the ATRA-mediated decrease in cholesterol content as well as increase in lysosomal acidification. In the context of disease, NPC2 expression is decreased in caseous tuberculosis granulomas and infected monocytes compared to normal lung tissue and uninfected cells, respectively. Stimulation of M. tuberculosis H37Ra-infected monocytes or M. tuberculosis H37Rv-infected MDMs with ATRA recovered NPC2 expression levels, and knockdown of NPC2 expression ablated the ATRA-induced antimicrobial activity, suggesting that NPC2 plays a pivotal role in vitamin A-mediated host defense. These results demonstrate that vitamin A-induced immune defense against M. tuberculosis is dependent on the expression and function of NPC2.
Regulation of cholesterol is an important facet of the host-pathogen interaction between immune cells and M. tuberculosis. In a caseous tuberculosis granuloma there is an increased expression of genes involved in lipid sequestration and metabolism as well as an accumulation of cholesterol, cholesteryl esters, triacylglycerols and lactosylceramide (40). The presence of foamy macrophages in the granuloma (42) and host hypercholesterolemia both correlate with loss of protection against M. tuberculosis (43). Studies have also demonstrated that phagocytosis of M. tuberculosis (27) and bacterial persistence within the macrophages are dependent on cholesterol (28). M. tuberculosis can accumulate and utilize cholesterol as a source of nutrition (29,44,45) as well as exploit host-derived lipids to reduce metabolic stress (46), which could be a determinant of pathogen virulence and immunogenicity (47). Accumulation of lipids within lysosomes alters the pH of the vesicle to favor bacterial survival (38) and fusion of lysosomes with phagosomes harboring M. tuberculosis is a critical host defense process against the infection (39,48,49).
Our data demonstrate that the ATRA-mediated decrease in cellular cholesterol requires the expression and function of NPC2. Mutations in the NPC2 gene have been well defined to be responsible for Niemann-Pick disease type C2, a lysosomal storage disease characterized by abnormally high cholesterol accumulation in cells. Our experiments show that loss of NPC2 expression not only ablated the ATRA-mediated reduction in cellular cholesterol content, but also resulted in a significant accumulation of cellular cholesterol, paralleling the cellular etiology of Niemann-Pick disease. Important to macrophage antimicrobial defense against M. tuberculosis, knockdown of NPC2 expression also ablated ATRA-induced lysosomal acidification, which is required for antimicrobial activity against the infection (38,39,48,49). Thus, decreasing NPC2 expression levels during infection can inhibit important macrophage defense mechanisms as well as increase a nutrient source within the cell, which favors bacterial survival. These data suggest that regulation of cellular cholesterol through proteins such as NPC2 may be a key part of the innate immune response and further studies investigating the consequence of cellular cholesterol modulation on bacterial viability are warranted.
Although vitamin A deficiency is associated with tuberculosis, vitamin A supplementation has not proven effective as a treatment (50,51). This is likely because vitamin A status is determined by serum retinol levels, and vitamin A supplementation similarly modulates retinol levels. In contrast, the ability to induce antimicrobial activity through vitamin A metabolites, such as in this and previous studies (3,4) utilize the active ATRA form. This suggests that while activation of infected macrophages with ATRA results in antimicrobial activity against intracellular M. tuberculosis infection, the intricate pathways that regulate retinol metabolism, especially in tuberculosis patients are still unclear. If retinol metabolism were inhibited by infection, then the effects of vitamin A supplementation will be effectively negated. Further studies are needed to understand how vitamin A is metabolized during the immune response to M. tuberculosis infection.
In summary, our data demonstrate a novel role for NPC2 in the ATRA-mediated innate immune response against M. tuberculosis, which suggests that regulation of intracellular cholesterol maybe an important facet of defense against infection. Understanding how vitamin A-mediated functions are regulated during infection in the host will be an important step in determining how this micronutrient can influence the outcome of disease.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. J.S. Adams, Dr. B.R. Bloom, and Dr. R.L. Modlin for critical reading of the manuscript. Nel and Rit were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Experiments using virulent M. tuberculosis were performed in the UCLA MIMG BSL-3 Select Agent Facility with approval and support from the Director of UCLA High Containment Facilities and the Office of the Vice Chancellor for Research.
GRANT SUPPORT This research is supported by a NIH K22 Career Development Award (AI 85025) awarded to PTL.
REFERENCES
- 1.Ramachandran G, Santha T, Garg R, Baskaran D, Iliayas SA, Venkatesan P, Fathima R, Narayanan PR. Vitamin A levels in sputum-positive pulmonary tuberculosis patients in comparison with household contacts and healthy `normals'. Int. J. Tuberc. Lung Dis. 2004;8:1130. [PubMed] [Google Scholar]
- 2.Mugusi FM, Rusizoka O, Habib N, Fawzi W. Vitamin A status of patients presenting with pulmonary tuberculosis and asymptomatic HIV-infected individuals, Dar es Salaam, Tanzania. Int. J. Tuberc. Lung Dis. 2003;7:804. [PubMed] [Google Scholar]
- 3.Crowle AJ, Ross EJ. Inhibition by retinoic acid of multiplication of virulent tubercle bacilli in cultured human macrophages. Infect. Immun. 1989;57:840. doi: 10.1128/iai.57.3.840-844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand PK, Kaul D, Sharma M. Synergistic action of vitamin D and retinoic acid restricts invasion of macrophages by pathogenic mycobacteria. J. Microbiol. Immunol. Infect. 2008;41:17. [PubMed] [Google Scholar]
- 5.Carlberg C. Lipid soluble vitamins in gene regulation. Biofactors. 1999;10:91. doi: 10.1002/biof.5520100202. [DOI] [PubMed] [Google Scholar]
- 6.Grange JM, Davies PD, Brown RC, Woodhead JS, Kardjito T. A study of vitamin D levels in Indonesian patients with untreated pulmonary tuberculosis. Tubercle. 1985;66:187. doi: 10.1016/0041-3879(85)90035-2. [DOI] [PubMed] [Google Scholar]
- 7.Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet. 2000;355:618. doi: 10.1016/S0140-6736(99)02301-6. [DOI] [PubMed] [Google Scholar]
- 8.Crowle AJ, Ross EJ, May MH. Inhibition by 1,25(OH)2-vitamin D3 of the multiplication of virulent tubercle bacilli in cultured human macrophages. Infect. Immun. 1987;55:2945. doi: 10.1128/iai.55.12.2945-2950.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, Kamen DL, Wagner M, Bals R, Steinmeyer A, Zugel U, Gallo RL, Eisenberg D, Hewison M, Hollis BW, Adams JS, Bloom BR, Modlin RL. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 10.Liu PT, Stenger S, Tang DH, Modlin RL. Cutting edge: vitamin D-mediated human antimicrobial activity against Mycobacterium tuberculosis is dependent on the induction of cathelicidin. J. Immunol. 2007;179:2060. doi: 10.4049/jimmunol.179.4.2060. [DOI] [PubMed] [Google Scholar]
- 11.Liu PT, Schenk M, Walker VP, Dempsey PW, Kanchanapoomi M, Wheelwright M, Vazirnia A, Zhang X, Steinmeyer A, Zugel U, Hollis BW, Cheng G, Modlin RL. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS. One. 2009;4:e5810. doi: 10.1371/journal.pone.0005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu PT, Krutzik SR, Kim J, Modlin RL. Cutting edge: all-trans retinoic acid down-regulates TLR2 expression and function. J. Immunol. 2005;174:2467. doi: 10.4049/jimmunol.174.5.2467. [DOI] [PubMed] [Google Scholar]
- 13.Liu PT, Phan J, Tang D, Kanchanapoomi M, Hall B, Krutzik SR, Kim J. CD209(+) macrophages mediate host defense against Propionibacterium acnes. J. Immunol. 2008;180:4919. doi: 10.4049/jimmunol.180.7.4919. [DOI] [PubMed] [Google Scholar]
- 14.Fabri M, Stenger S, Shin DM, Yuk JM, Liu PT, Realegeno S, Lee HM, Krutzik SR, Schenk M, Sieling PA, Teles R, Montoya D, Iyer SS, Bruns H, Lewinsohn DM, Hollis BW, Hewison M, Adams JS, Steinmeyer A, Zugel U, Cheng G, Jo EK, Bloom BR, Modlin RL. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci. Transl. Med. 2011;3:104ra102. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu PT, Wheelwright M, Teles R, Komisopoulou E, Edfeldt K, Ferguson B, Mehta MD, Vazirnia A, Rea TH, Sarno EN, Graeber TG, Modlin RL. MicroRNA-21 targets the vitamin D-dependent antimicrobial pathway in leprosy. Nat. Med. 2012:10. doi: 10.1038/nm.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, Moraes MO, Williams DL. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J. Clin. Microbiol. 2009;47:2124. doi: 10.1128/JCM.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S, Fan G. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500:593. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szeles L, Poliska S, Nagy G, Szatmari I, Szanto A, Pap A, Lindstedt M, Santegoets SJ, Ruhl R, Dezso B, Nagy L. Research resource: transcriptome profiling of genes regulated by RXR and its permissive and nonpermissive partners in differentiating monocyte-derived dendritic cells. Mol. Endocrinol. 2010;24:2218. doi: 10.1210/me.2010-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szeles L, Keresztes G, Torocsik D, Balajthy Z, Krenacs L, Poliska S, Steinmeyer A, Zuegel U, Pruenster M, Rot A, Nagy L. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J. Immunol. 2009;182:2074. doi: 10.4049/jimmunol.0803345. [DOI] [PubMed] [Google Scholar]
- 22.Zolfaghari R, Chen Q, Ross AC. DHRS3, a retinal reductase, is differentially regulated by retinoic acid and lipopolysaccharide-induced inflammation in THP-1 cells and rat liver. Am. J. Physiol Gastrointest. Liver Physiol. 2012;303:G578–G588. doi: 10.1152/ajpgi.00234.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS. Comput. Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. %20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Mellor J, Wu J, Yamada T, Holloway D, Delisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetti A, Lelong E, Cosson P. A measure of endosomal pH by flow cytometry in Dictyostelium. BMC. Res. Notes. 2009;2:7. doi: 10.1186/1756-0500-2-7. doi: 10.1186/1756-0500-2-7.:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmann T, Liebisch G, Moehle C, Schifferer R, Dayoub R, Heiduczek S, Grandl M, Dada A, Schmitz G. Gene expression profiling identifies retinoids as potent inducers of macrophage lipid efflux. Biochim. Biophys. Acta. 2005;1740:155. doi: 10.1016/j.bbadis.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 27.Gatfield J, Pieters J. Essential role for cholesterol in entry of mycobacteria into macrophages. Science. 2000;288:1647. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen L, Pieters J. The Trojan horse: survival tactics of pathogenic mycobacteria in macrophages. Trends Cell Biol. 2005;15:269. doi: 10.1016/j.tcb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Brzostek A, Pawelczyk J, Rumijowska-Galewicz A, Dziadek B, Dziadek J. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 2009;191:6584. doi: 10.1128/JB.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costet P, Lalanne F, Gerbod-Giannone MC, Molina JR, Fu X, Lund EG, Gudas LJ, Tall AR. Retinoic acid receptor-mediated induction of ABCA1 in macrophages. Mol. Cell Biol. 2003;23:7756. doi: 10.1128/MCB.23.21.7756-7766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escher G, Krozowski Z, Croft KD, Sviridov D. Expression of sterol 27-hydroxylase (CYP27A1) enhances cholesterol efflux. J. Biol. Chem. 2003;278:11015. doi: 10.1074/jbc.M212780200. [DOI] [PubMed] [Google Scholar]
- 32.Mukhamedova N, Rose H, Cui HL, Grant A, Tchoua U, Dart A, Bukrinsky M, Sviridov D. Antiretroviral compounds and cholesterol efflux from macrophages. Atherosclerosis. 2009;206:439. doi: 10.1016/j.atherosclerosis.2009.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Mu H, Chai H, Liao D, Yao Q, Chen C. Human immunodeficiency virus protease inhibitor ritonavir inhibits cholesterol efflux from human macrophage-derived foam cells. Am. J. Pathol. 2007;171:304. doi: 10.2353/ajpath.2007.060965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC. Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. doi: 10.1186/1471-2105-9-559.:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixit SS, Jadot M, Sohar I, Sleat DE, Stock AM, Lobel P. Loss of Niemann-Pick C1 or C2 protein results in similar biochemical changes suggesting that these proteins function in a common lysosomal pathway. PLoS. One. 2011;6:e23677. doi: 10.1371/journal.pone.0023677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman SD, Krise JP. Niemann-Pick C1 functions independently of Niemann-Pick C2 in the initial stage of retrograde transport of membrane-impermeable lysosomal cargo. J. Biol. Chem. 2010;285:4983. doi: 10.1074/jbc.M109.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boadu E, Nelson RC, Francis GA. ABCA1-dependent mobilization of lysosomal cholesterol requires functional Niemann-Pick C2 but not Niemann-Pick C1 protein. Biochim. Biophys. Acta. 2012;1821:396. doi: 10.1016/j.bbalip.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 38.Cox BE, Griffin EE, Ullery JC, Jerome WG. Effects of cellular cholesterol loading on macrophage foam cell lysosome acidification. J. Lipid Res. 2007;48:1012. doi: 10.1194/jlr.M600390-JLR200. [DOI] [PubMed] [Google Scholar]
- 39.Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, Allen RD, Gluck SL, Heuser J, Russell DG. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 40.Kim MJ, Wainwright HC, Locketz M, Bekker LG, Walther GB, Dittrich C, Visser A, Wang W, Hsu FF, Wiehart U, Tsenova L, Kaplan G, Russell DG. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2010;2:258. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Broser M, Rom WN. Activation of the interleukin 6 gene by Mycobacterium tuberculosis or lipopolysaccharide is mediated by nuclear factors NF-IL6 and NF-kappa B. Proc. Natl. Acad. Sci. U. S. A. 1994;91:2225. doi: 10.1073/pnas.91.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009;10:943. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martens GW, Arikan MC, Lee J, Ren F, Vallerskog T, Kornfeld H. Hypercholesterolemia impairs immunity to tuberculosis. Infect. Immun. 2008;76:3464. doi: 10.1128/IAI.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS. Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 2012;19:218. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee W, Vanderven BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis Exploits Host-derived Fatty Acids to Limit Metabolic Stress. J. Biol. Chem. 2013;288:6788. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koo MS, Subbian S, Kaplan G. Strain specific transcriptional response in Mycobacterium tuberculosis infected macrophages. Cell Commun. Signal. 2012;10:2. doi: 10.1186/1478-811X-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4033. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welin A, Raffetseder J, Eklund D, Stendahl O, Lerm M. Importance of phagosomal functionality for growth restriction of Mycobacterium tuberculosis in primary human macrophages. J. Innate. Immun. 2011;3:508. doi: 10.1159/000325297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pakasi TA, Karyadi E, Suratih NM, Salean M, Darmawidjaja N, Bor H, d. van V, Dolmans WM, Van der Meer JW. Zinc and vitamin A supplementation fails to reduce sputum conversion time in severely malnourished pulmonary tuberculosis patients in Indonesia. Nutr. J. 2010;9:41. doi: 10.1186/1475-2891-9-41. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visser ME, Grewal HM, Swart EC, Dhansay MA, Walzl G, Swanevelder S, Lombard C, Maartens G. The effect of vitamin A and zinc supplementation on treatment outcomes in pulmonary tuberculosis: a randomized controlled trial. Am. J. Clin. Nutr. 2011;93:93. doi: 10.3945/ajcn.110.001784. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.