Fig. 2. Binding analysis of phagotopes and peptides to UN1 mAb.
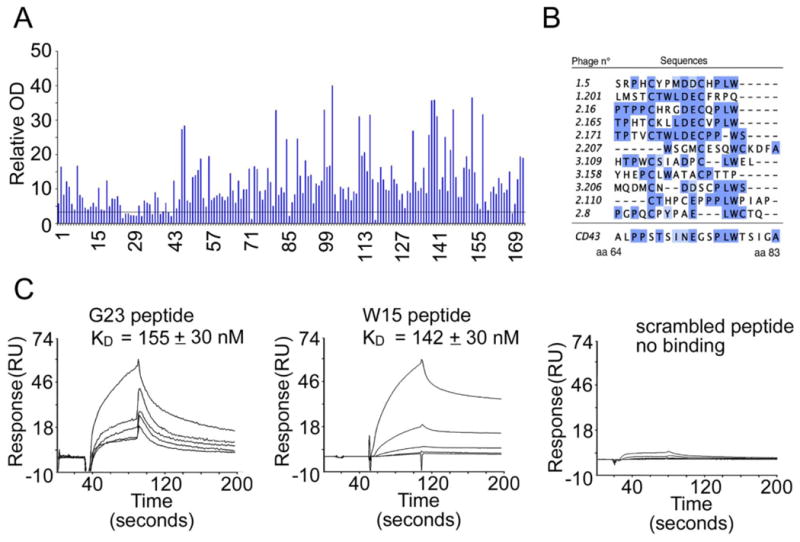
(A) ELISA binding analysis of 174 selected phages clones to UN1 mAb. Each phagotope was tested in duplicate and the relative absorbance was calculated as the difference between OD405nm and OD620nm. Relative OD indicates the ratio of the mean OD value of the tested phagotope to the mean OD value of the wild type phage.
(B) MUSCLE-based alignment of peptide sequence of selected phagotopes with CD43 protein. The positions of amino acids in CD43 aligned sequence are indicated.
(C) Affinity measurement of G23 and W15 peptides binding to UN1 mAb by Surface Plasmon Resonance (SPR). The sensorgrams show G23 peptide, W15 peptide and scrambled peptide binding to UN1 mAb immobilized on CM5 sensor chip. The peptide concentrations used were 0.1, 0.25, 0.5, 1.0 and 2.5 mM. The response expressed in resonance units was recorded as a function of time. For G23 and W15 peptide, the corresponding KD is indicated.
