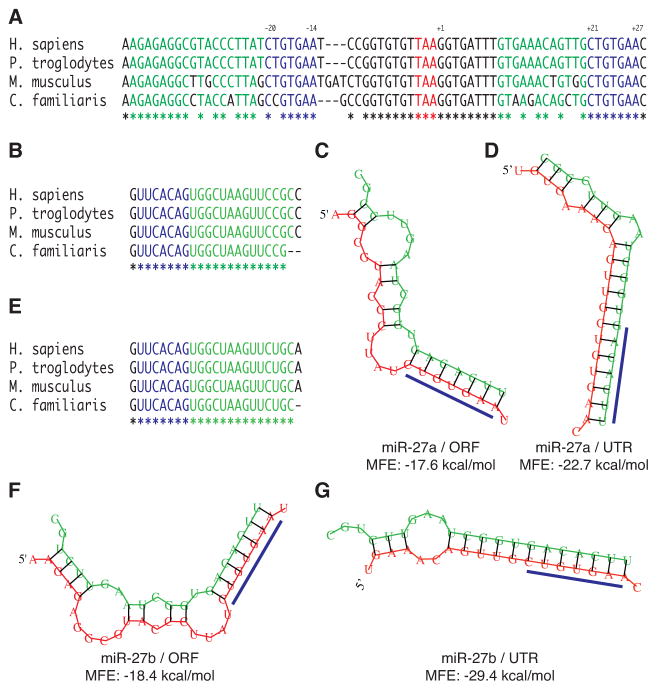
Figure 1. Identification of candidate miR-27a and miR-27b binding sites in the DPYD gene.
A, alignment of mammalian DPYD (NM_000110) sequences for Homo sapiens, Pan troglodytes, Mus musculus, and Canis familiaris. The stop codon is indicated by red lettering. Predicted seed binding sequences are indicated by blue lettering, and predicted flanking microRNA binding sequences are indicated by green lettering. Fully conserved nucleotides are indicated by an asterisk below the alignment. The position of the predicted seed binding regions is reported above the alignment relative to the first nucleotide of the 3′ UTR with coordinates reported relative to the Homo sapiens sequence. B, conservation of mature miR-27a sequences. The minimum free energy (MFE) structures of miR-27a complexed to the predicted ORF (C) and UTR (D) binding sites of Homo sapiens DPYD were predicted using RNAhybrid (25). E, sequence conservation of mature miR-27b. Predicted structures of miR-27b bound to the ORF (F) and UTR (G) binding sites.