Fig. 2. Structures of gp120 V1/V2/V3 variable loops.
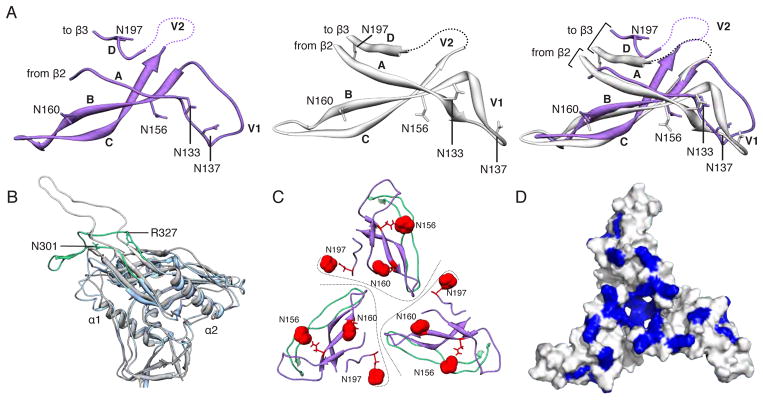
(A) Comparison of the V1/V2 domain from the EM model (purple) and the previously published X-ray structure (3U2S, white) with key residues labeled. The structures are nearly identical except for the base of V1/V2 (strands A and D) and the V1 loop. (B) Superposition of gp120 and the V3 loop from the EM model (blue/green) onto a previous X-ray structure containing a V3 loop (2B4C, gray/white). While gp120 is structurally conserved, the V3 β-hairpin loop exhibits a different configuration. Specifically, V3 bends around a hinge formed near residues 302 and 326. The N-terminal residues of V3 up to residue 301 and the C-terminal residues after residue 327 are structurally similar. Residues 1–90, and 126–196 are omitted for clarity. (C) Quaternary arrangement of V1, V2, and V3 regions of gp120 at the top of the trimer. The positions of glycans at N156, N160, and N197 have been highlighted as red spheres. Lines denote approximate protomer boundaries. (D) Surface view of the modeled portion in C, with the basic residues (Arg and Lys) colored blue. A large number of basic amino acids are localized to the trimer apex.
