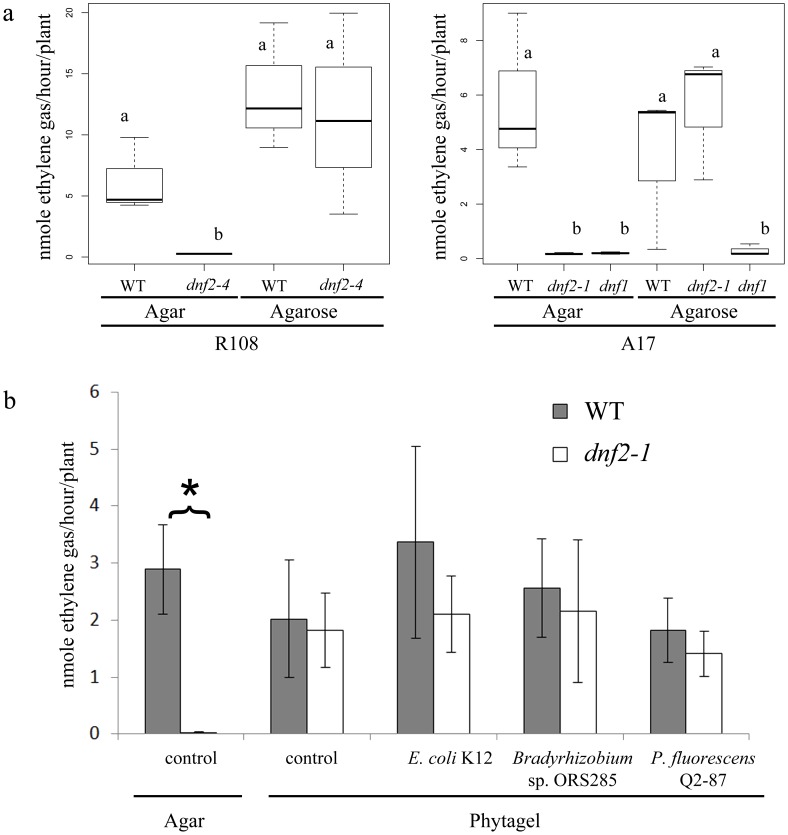
Figure 3. Plant growth conditions determine the DNF2 requirement for symbiosis.
a) Acetylene reduction assays were conducted on M. truncatula WT ecotype R108 and A17 and dnf2–4 (R108 genetic background), and dnf2-1, dnf1 mutants (A17 genetic background) inoculated with S. medicae strain WSM419. A Kruskal-Wallis one-way ANOVA test and a post-hoc Tukey’s test were performed and statistically identical values were attributed identical letters (n = 3). (b) Acetylene reduction assay was conducted on M. truncatula WT ecotype A17 and dnf2-1 mutant inoculated with strain WSM419 alone (control) or in combination with E. coli K12, Bradyrhizobium sp. ORS285 or P. fluorescens Q2–87. Plants were grown under in vitro conditions on BNM supplemented with either agar or agarose and analyzed at 14 dpi (a) or with Agar and Phytagel and analyzed at 25 dpi (b). A Mann-Whitney test was performed between WT and dnf2-1 mutant for each condition. The star indicates a significant difference (p-value <0.05) (n = 4) Error bars represent standard errors.