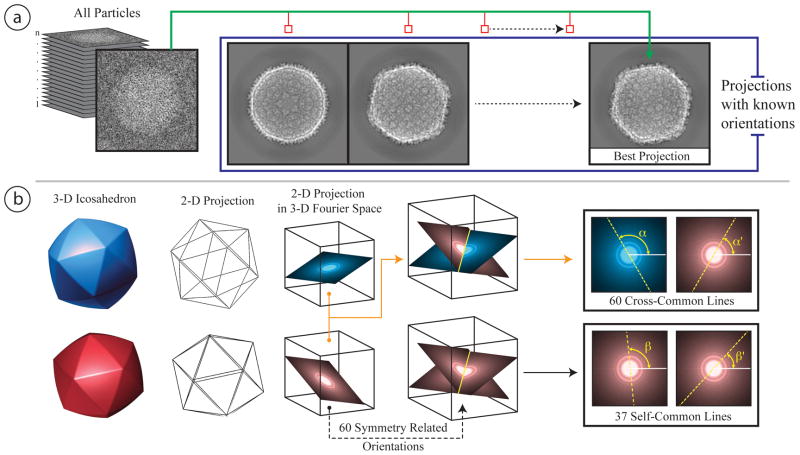
Figure 4.
Single particle 2D alignment methods. (a) Schematic of projection matching method. By comparing individual raw single particle images to projections made of a model at known orientations, it is possible to define classes of particles that can then be averaged together to obtain a 3-D electron density map. (b) Cross-common line method. According to central section theorem, the Fourier transforms of any two images/projections intersect along a line where the values are identical. If the two images are generated from two totally different orientations, the intersection line is called cross common line (top of figure 4b). Considering the icosahedral symmetry of a virus particle, there are 60 pairs of cross common lines between the two images. If the orientations of the two images are only related to its icosahedral symmetry, they are actually the same image. The intersection line is called self-common line. There are 37 unique pairs of self-common lines for an icosahedral particle image.