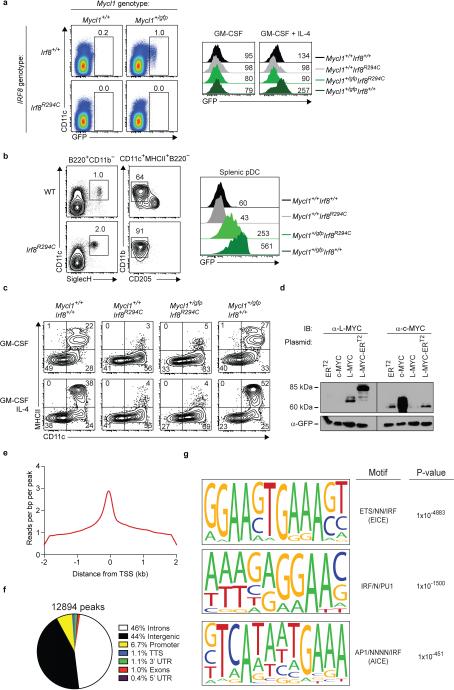
Extended Data Figure 5. IRF8 regulates the expression of Mycl1 in multiple dendritic cell lineages.
a, (left panels) BM cells and splenocytes from Mycl1+/+Irf8+/+, Mycl1+/gfpIrf8+/+, Mycl1+/+Irf8R294C, and Mycl1+/gfpIrf8R294C mice were stained for analysis. Shown are two-color histograms of GFP and CD11c expression. Numbers represent the percent of cells in the indicate gates. Data are representative of 3 independent experiments. (right panels) Ly6C+CD11b+ BM monocytes were isolated by cell sorting from Mycl1+/+Irf8+/+(black line), Mycl1+/+Irf8R294C(grey line), Mycl1+/gfpIrf8R294C(light green line), Mycl1+/gfpIrf8+/+mice (dark green line) and differentiated for 4 days either with GM-CSF (left panel) or with GM-CSF and IL-4 (right panel). Shown are single-color histograms of GFP expression for pre-gated live CD11c+ cells from the indicated genotypes. b, Splenocytes from WT (top panels) and Irf8R294C (bottom panels) mice on the C57BL/6 genetic background were stained for analysis. Shown are two-color histograms of CD11c and SiglecH expression (left panels), and CD11b and CD205 expression (right panels), for cells pre-gated as either B220+CD11b− or CD11c+MHCII+B220−. pDCs were gated as CD11c+SiglecH+B220+CD11b−. Numbers represent the percent of cells in the indicated gates. Data are representative of 2 independent experiments. c, Ly6C+ BM monocytes were isolated by cell sorting from Mycl1+/+Irf8+/+, Mycl1+/+Irf8R294C, Mycl1+/gfpIrf8R294C, and Mycl1+/gfpIrf8+/+ mice. Monocytes were cultured either with GM-CSF or with GM-CSF and IL-4 for 4 days and then stained for analysis. Shown are two-color histograms of MHCII and CD11c expression for differentiated monocytes. Numbers represent the percent of cells in the quadrant gate. Data are representative of 2 independent experiments. d, Phoenix-E cells were transfected with murine stem cell virus (MSCV) retroviral plasmids expressing the mutant human estrogen receptor (ERT2), murine Myc (c-MYC), murine Mycl1 (L-MYC), and a fusion between Mycl1 and the mutant human estrogen receptor (L-MYC-ERT2). Whole-cell extracts were prepared 2 days after transfection and Western blot analysis was performed. Shown are blots probed for L-MYC (left, α-L-MYC) and c-MYC (right, α-c-MYC) for the indicated transfections. Blots were stripped and re-probed for GFP (bottom, α-GFP). e, IRF8 DNA binding was assayed by ChIP-Seq. Shown is a histogram of normalized reads per bp per peak within a 4-kb window centered on transcriptional start sites (TSS). f, Uniquely mapped IRF8 ChIP-Seq reads were evaluated for peak discovery by Model-based Analysis of ChIP-Seq (MACS) as described in Methods and peaks were annotated by the Homer software package using default annotation settings. Shown is the distribution of peak annotations within introns, intergenic regions, promoters, transcription termination sites (TTS), 3'-untranslated regions (3'UTR), exons, and 5'-untranslated regions (5'UTR). Numbers represent the percent of annotations in each category. g, Peaks identified in (f) were evaluated for de novo motif discovery using the Homer software package as described in Methods. Analysis was performed 3 separate times to identify the most significant motifs. Shown are the top 3 DNA motifs conserved in IRF8-bound loci along with the closest known motif and associated p-value (cumulative binomial distribution).