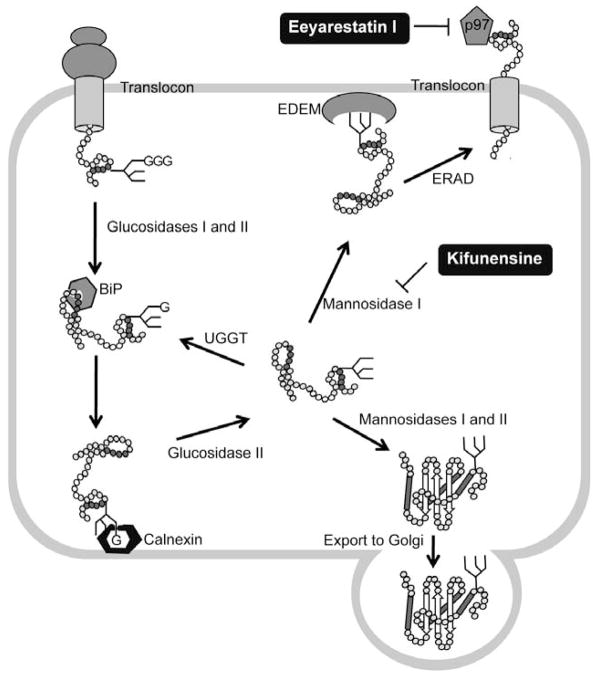
Fig. 4.
ER-associated degradation (ERAD) and its chemical modulators. Proteins entering the endoplasmic reticulum (ER) are immediately recognized by BiP and often modified by the addition of a GlcNAc2-Man9-Glc3 glycan. Glucosidases I and II sequentially remove two terminal glucoses (G) from the glycan and generate monoglucosylated substrates that are recognized by calnexin and calreticulin (calreticulin is a soluble protein and is not shown), which facilitate substrate folding. Once the substrate is released from the calnexin–calreticulin cycle, glucosidase II trims the last glucose. Proteins that have adopted their native conformation are demannosylated by mannosidases I and II and exit the ER. If proteins have not been folded properly, they re-enter the calnexin–calreticulin cycle. Such proteins are reglucosylated by UDP-glucose:glycoprotein glucosyltransferase (UGGT), which promotes re-entry into the folding cycle. Terminally misfolded proteins are processed by mannosidase I and then targeted for ERAD with the participation, through an undetermined mechanism, of the ER degradation-enhancing -mannosidase-like lectins (EDEM). Retrotranslocation of the misfolding-prone substrate to the cytoplasm is mediated by p97 complex. The small molecule ERAD inhibitors kifunesine and Eeyarestatin I block different steps of the ERAD pathways.