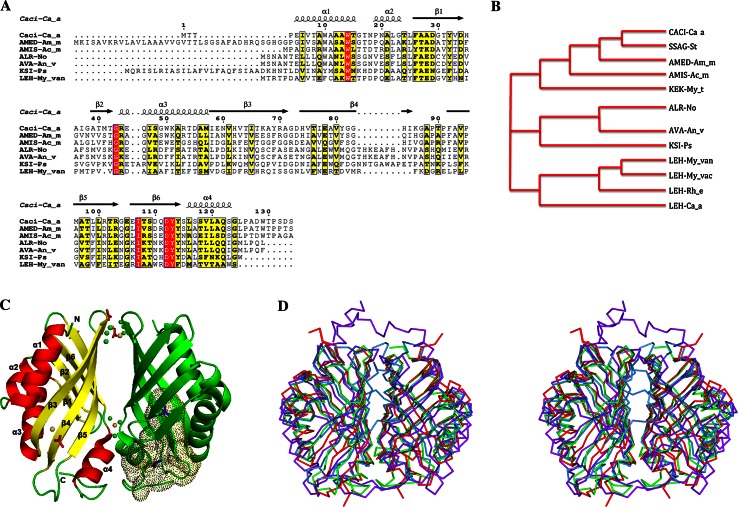
Fig. 1.
Sequence alignment and Overall structure of the Caci_0382 protein. a and b Sequence alignment and phylogenetic tree of Caci_0382 from Catenulispora acidiphila DSM 44928 (Caci-Ca_a) and its homologs: SSAG_04049 from Streptomyces sp. Mg1 (SSAG_St), AMED_3668 from Amycolatopsis mediterranei U32 (AMED-Am_m), AMIS_27750 from Actinoplanes missouriensis 431 (AMIS-Ac_m), ALR3729 from Nostoc sp. PCC 7120 (ALR-No), Ava_1595 from Anabaena variabilis ATCC 29413 (AVA-An_v), KSI-like protein from Pseudomonas sp. GM78 (KSI-Ps), KEK_02356 from Mycobacterium thermoresistibile ATCC 19527 (KEK-My_t), LEH from Mycobacterium vanbaalenii PYR-1 (LEH-My_van), LEH from Mycobacterium vaccae ATCC 25954 (LEH-My_vac), LEH from Rhodococcus erythropolis (LEH-Rh_e), LEH from Catenulispora acidiphila DSM 44928 (LEH-Ca_a). Secondary structure elements of Caci_0382 are indicated above the sequence. Sequence homologies are highlighted by red background (identities) and yellow (similarities). c Ribbon diagram of the Caci_0382 dimer in complex with TMA (shown as the stick blue model in one subunit). Cadmium (yellow), chloride (green) and acetate (red) ions are shown as ball and stick model, respectively. The binding cavity in one subunit of the Caci_0382 structure is presented as dots. The secondary structure elements of the one subunit are labeled. d Stereo view of the structural homologs [LEH (light blue), NTF2 (red), and PHZB (purple)] superposed on the Caci_0382 dimeric structure (green)