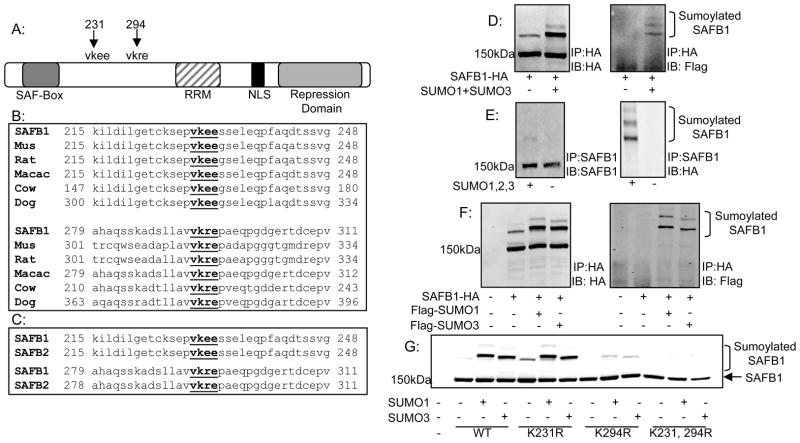
Figure 1. Identification and conservation of SAFB1 sumoylation sites.
A: Schematic of SAFB1 showing major domains and lysine’s at amino acid positions 231 and 294 which conform to the consensus sumoylation sequence. B: Alignment of region containing SAFB1 sumoylation sites using the ClustalW web tool indicating conservation of the sumoylation sites with lower organisms. C: Alignment as described in B indicating conservation of SAFB1 sumoylation sites in paralog SAFB2. D: 293T cells were transfected with constructs for Flag-SUMO1 and Flag-SUMO3 with the addition of either SAFB1-HA or SAFB2-GFP. Cells were collected and immunoprecipitated for either HA or GFP as indicated. Immunoblotting for HA or GFP and FLAG revealed two sumoylated SAFB1 bands. E: Performed as in D but with IP of endogenous SAFB1. F: 293T cells were transfected with SAFB1-HA and Flag-SUMO1 or Flag-SUMO3. Cells were collected and immunoprecipitated for HA. Immunoblotting for Flag revealed sumoylation of SAFB1 by both SUMO1 and SUMO3. G: 293T cells were transfected with varying SAFB1 constructs and my-SUMO1 or SUMO3 as indicated. Cells were collected and immunoblotted for HA. Results indicate complete loss of sumoylated forms with mutation of lysine’s 231 and 294.