Figure 2.
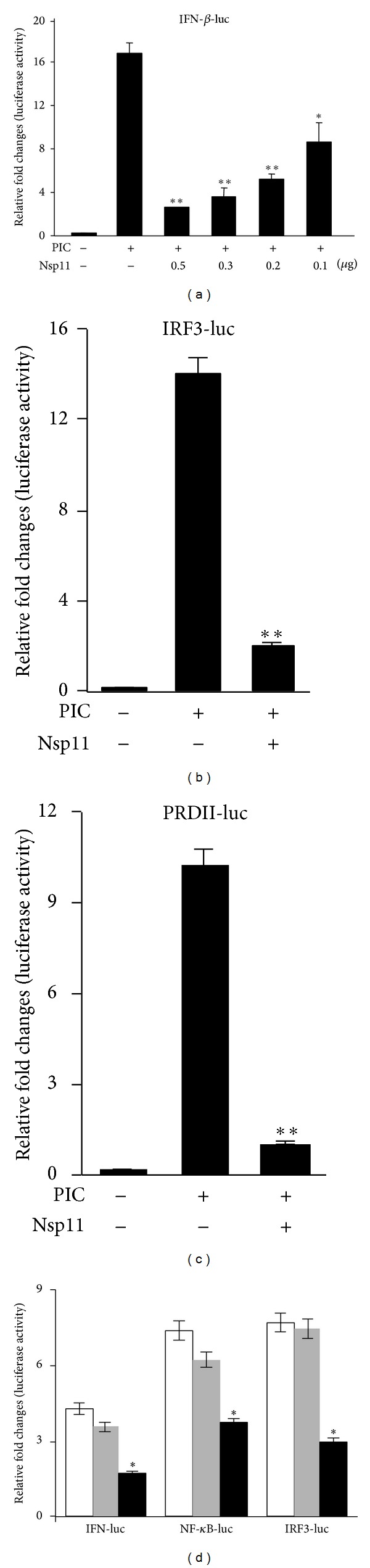
Suppression of type I IFN induction by PRRSV nsp11 in gene-transfected MARC-145 cells (a, b, and c), and stably-expressing MARC-nsp11 cells (d). (a) MARC-145 cells were seeded in 12-well plates and transfected with pXJ41 (0.5 μg) or pXJ41-Flag-nsp11 plasmid of 0.5, 0.3, 0.2, or 0.1 μg, together with pRL-TK (0.05 μg) and pIFN-β-luc (0.5 μg) reporter. After 24 h, cells were transfected with 0.5 μg/mL of poly (I:C) for 16 h and harvested for luciferase assay (Promega). (b) and (c) MARC-145 cells were cotransfected with either pIRF3-luc (0.5 μg) or pPRDII-luc (0.5 μg) or pXJ41-Flag-nsp11 (0.5 μg) or pXJ41 (0.5 μg), along with pRL-TK. After 24 h, cells were transfected with 0.5 μg/mL of poly (I:C) for 16 h and harvested for luciferase assay (Promega). The experiments were conducted in duplicate and repeated three times, and the overage values were depicted. The fold change was calculated by (PIC+)/(PIC−) for each sample. The negative control represents the basal level luciferase activity. The values from nsp11 gene-transfected samples were compared with those of poly (I:C) stimulation, and the data were analyzed using 2-tail t-test. One star (∗) represents P < 0.01 and two stars (∗∗) represent P < 0.005. (d) MARC-145 or MARC-nsp11 cells were cotransfected with pIFN-β-luc, pIRF3-luc, or pPRDII-luc and pRL-TK. MARC-145 cells were transfected with the pLNCX2 empty vector as a negative control. All samples were treated and processed as described above. Luciferase assays were conducted in duplicate and repeated three times. One star (∗) represents P < 0.05. White bars represent MARC-145 cells, grey bars represent the pLNCX2 retrovirus expression vector-transfected MARC-145 cells, and black bars represent MARC-nsp11 cells.
