Abstract
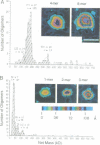
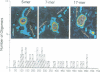
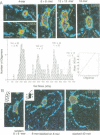
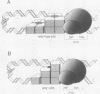
We examined the role of p53 oligomerization in DNA binding and in transactivation. By conventional electron microscopy (EM) and scanning transmission EM, we find that wild-type tetramers contact 18-20 bp at single or tandem 19 bp consensus sequences and also stack in apparent register, tetramer on top of tetramer. Stacked tetramers link separated DNA binding sites with DNA loops. Interestingly, the p53(1-320) segment, which lacks the C-terminal tetramerization domain, binds DNA consensus sites as stacked oligomers. Although the truncated protein binds DNA with reduced efficiency, it nevertheless induces DNA looping by self-association. p53, therefore, has a C-terminal tetramerization domain that enhances DNA binding and a non-tetrameric oligomerization domain that stacks p53 at consensus sites and loops separated consensus sites via protein-protein interactions. Using model promoters, we demonstrate that wild-type and tetramerization-deficient p53s activate transcription well when tandem consensus sites are proximal to TATA sequences and poorly when tandem sites are distal. In the presence of proximal sites, however, stimulation by distal sites increases 25-fold. Tetramerization and stacking of tetramers, therefore, provide dual mechanisms to augment the number of p53 molecules available for activation through p53 response elements. DNA looping between separated response elements further increases the concentration of local p53 by translocating distally bound protein to the promoter.
Full text
PDF


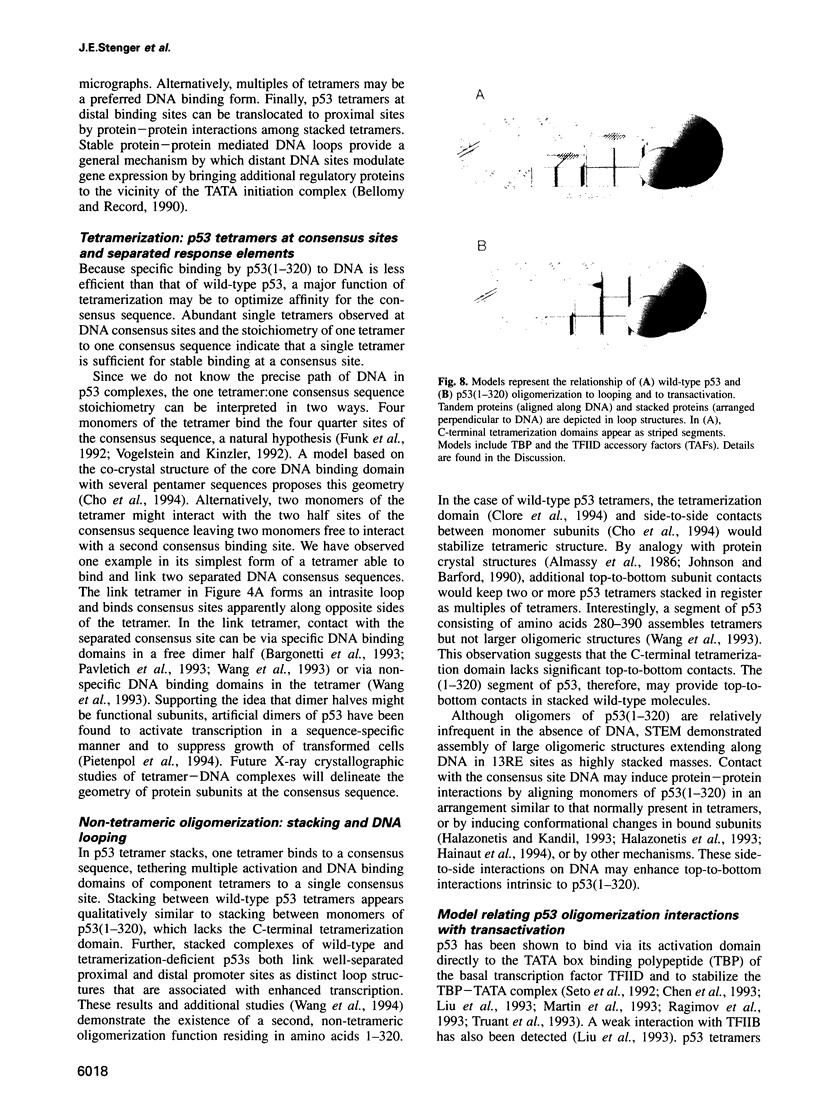


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almassy R. J., Janson C. A., Hamlin R., Xuong N. H., Eisenberg D. Novel subunit-subunit interactions in the structure of glutamine synthetase. 1986 Sep 25-Oct 1Nature. 323(6086):304–309. doi: 10.1038/323304a0. [DOI] [PubMed] [Google Scholar]
- Bargonetti J., Friedman P. N., Kern S. E., Vogelstein B., Prives C. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell. 1991 Jun 14;65(6):1083–1091. doi: 10.1016/0092-8674(91)90560-l. [DOI] [PubMed] [Google Scholar]
- Bargonetti J., Manfredi J. J., Chen X., Marshak D. R., Prives C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993 Dec;7(12B):2565–2574. doi: 10.1101/gad.7.12b.2565. [DOI] [PubMed] [Google Scholar]
- Beachy P. A., Varkey J., Young K. E., von Kessler D. P., Sun B. I., Ekker S. C. Cooperative binding of an Ultrabithorax homeodomain protein to nearby and distant DNA sites. Mol Cell Biol. 1993 Nov;13(11):6941–6956. doi: 10.1128/mcb.13.11.6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomy G. R., Record M. T., Jr Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog Nucleic Acid Res Mol Biol. 1990;39:81–128. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- Chen X., Farmer G., Zhu H., Prywes R., Prives C. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 1993 Oct;7(10):1837–1849. doi: 10.1101/gad.7.10.1837. [DOI] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P. D., Pavletich N. P. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994 Jul 15;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Omichinski J. G., Sakaguchi K., Zambrano N., Sakamoto H., Appella E., Gronenborn A. M. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science. 1994 Jul 15;265(5170):386–391. doi: 10.1126/science.8023159. [DOI] [PubMed] [Google Scholar]
- Farmer G., Bargonetti J., Zhu H., Friedman P., Prywes R., Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992 Jul 2;358(6381):83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- Fields S., Jang S. K. Presence of a potent transcription activating sequence in the p53 protein. Science. 1990 Aug 31;249(4972):1046–1049. doi: 10.1126/science.2144363. [DOI] [PubMed] [Google Scholar]
- Foord O. S., Bhattacharya P., Reich Z., Rotter V. A DNA binding domain is contained in the C-terminus of wild type p53 protein. Nucleic Acids Res. 1991 Oct 11;19(19):5191–5198. doi: 10.1093/nar/19.19.5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman P. N., Chen X., Bargonetti J., Prives C. The p53 protein is an unusually shaped tetramer that binds directly to DNA. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3319–3323. doi: 10.1073/pnas.90.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk W. D., Pak D. T., Karas R. H., Wright W. E., Shay J. W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992 Jun;12(6):2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg D., Mechta F., Yaniv M., Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P., Hall A., Milner J. Analysis of p53 quaternary structure in relation to sequence-specific DNA binding. Oncogene. 1994 Jan;9(1):299–303. [PubMed] [Google Scholar]
- Halazonetis T. D., Davis L. J., Kandil A. N. Wild-type p53 adopts a 'mutant'-like conformation when bound to DNA. EMBO J. 1993 Mar;12(3):1021–1028. doi: 10.1002/j.1460-2075.1993.tb05743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Kandil A. N. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 1993 Dec 15;12(13):5057–5064. doi: 10.1002/j.1460-2075.1993.tb06199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Hori R., Carey M. The role of activators in assembly of RNA polymerase II transcription complexes. Curr Opin Genet Dev. 1994 Apr;4(2):236–244. doi: 10.1016/s0959-437x(05)80050-4. [DOI] [PubMed] [Google Scholar]
- Hough P. V., Mastrangelo I. A., Wall J. S., Hainfeld J. F., Simon M. N., Manley J. L. DNA-protein complexes spread on N2-discharged carbon film and characterized by molecular weight and its projected distribution. J Mol Biol. 1982 Sep 15;160(2):375–386. doi: 10.1016/0022-2836(82)90183-8. [DOI] [PubMed] [Google Scholar]
- Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. Regulation of the specific DNA binding function of p53. Cell. 1992 Nov 27;71(5):875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Johnson L. N., Barford D. Glycogen phosphorylase. The structural basis of the allosteric response and comparison with other allosteric proteins. J Biol Chem. 1990 Feb 15;265(5):2409–2412. [PubMed] [Google Scholar]
- Kastan M. B., Zhan Q., el-Deiry W. S., Carrier F., Jacks T., Walsh W. V., Plunkett B. S., Vogelstein B., Fornace A. J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992 Nov 13;71(4):587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Kern S. E., Kinzler K. W., Baker S. J., Nigro J. M., Rotter V., Levine A. J., Friedman P., Prives C., Vogelstein B. Mutant p53 proteins bind DNA abnormally in vitro. Oncogene. 1991 Jan;6(1):131–136. [PubMed] [Google Scholar]
- Kern S. E., Kinzler K. W., Bruskin A., Jarosz D., Friedman P., Prives C., Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991 Jun 21;252(5013):1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- Kern S. E., Pietenpol J. A., Thiagalingam S., Seymour A., Kinzler K. W., Vogelstein B. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science. 1992 May 8;256(5058):827–830. doi: 10.1126/science.1589764. [DOI] [PubMed] [Google Scholar]
- Knight J. D., Li R., Botchan M. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Knight J. D., Jackson S. P., Tjian R., Botchan M. R. Direct interaction between Sp1 and the BPV enhancer E2 protein mediates synergistic activation of transcription. Cell. 1991 May 3;65(3):493–505. doi: 10.1016/0092-8674(91)90467-d. [DOI] [PubMed] [Google Scholar]
- Liu X., Miller C. W., Koeffler P. H., Berk A. J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993 Jun;13(6):3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D. H., Vartikar J., Pipas J. M., Laimins L. A. Specific repression of TATA-mediated but not initiator-mediated transcription by wild-type p53. Nature. 1993 May 20;363(6426):281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- Martin D. W., Muñoz R. M., Subler M. A., Deb S. p53 binds to the TATA-binding protein-TATA complex. J Biol Chem. 1993 Jun 25;268(18):13062–13067. [PubMed] [Google Scholar]
- Mastrangelo I. A., Courey A. J., Wall J. S., Jackson S. P., Hough P. V. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo I. A., Held P. G., Dailey L., Wall J. S., Hough P. V., Heintz N., Heintz N. H. RIP60 dimers and multiples of dimers assemble link structures at an origin of bidirectional replication in the dihydrofolate reductase amplicon of Chinese hamster ovary cells. J Mol Biol. 1993 Aug 5;232(3):766–778. doi: 10.1006/jmbi.1993.1430. [DOI] [PubMed] [Google Scholar]
- Mastrangelo I. A., Hough P. V., Wall J. S., Dodson M., Dean F. B., Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature. 1989 Apr 20;338(6217):658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Pascal E., Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991 Sep;5(9):1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Chambers K. A., Pabo C. O. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993 Dec;7(12B):2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- Pietenpol J. A., Tokino T., Thiagalingam S., el-Deiry W. S., Kinzler K. W., Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci U S A. 1994 Mar 15;91(6):1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M., Gann A. A. Activators and targets. Nature. 1990 Jul 26;346(6282):329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ragimov N., Krauskopf A., Navot N., Rotter V., Oren M., Aloni Y. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene. 1993 May;8(5):1183–1193. [PubMed] [Google Scholar]
- Raycroft L., Wu H. Y., Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990 Aug 31;249(4972):1049–1051. doi: 10.1126/science.2144364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed M., Wang Y., Mayr G., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: suppression, transformation, and transactivation. Gene Expr. 1993;3(1):95–107. [PMC free article] [PubMed] [Google Scholar]
- Schärer E., Iggo R. Mammalian p53 can function as a transcription factor in yeast. Nucleic Acids Res. 1992 Apr 11;20(7):1539–1545. doi: 10.1093/nar/20.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E., Usheva A., Zambetti G. P., Momand J., Horikoshi N., Weinmann R., Levine A. J., Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E., Zauberman A., Milner J., Davies E. A., Oren M. Tight DNA binding and oligomerization are dispensable for the ability of p53 to transactivate target genes and suppress transformation. EMBO J. 1993 Jul;12(7):2789–2797. doi: 10.1002/j.1460-2075.1993.tb05940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Langowski J., Baldwin R. L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger J. E., Mayr G. A., Mann K., Tegtmeyer P. Formation of stable p53 homotetramers and multiples of tetramers. Mol Carcinog. 1992;5(2):102–106. doi: 10.1002/mc.2940050204. [DOI] [PubMed] [Google Scholar]
- Stürzbecher H. W., Brain R., Addison C., Rudge K., Remm M., Grimaldi M., Keenan E., Jenkins J. R. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992 Aug;7(8):1513–1523. [PubMed] [Google Scholar]
- Su W., Jackson S., Tjian R., Echols H. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 1991 May;5(5):820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- Tarunina M., Jenkins J. R. Human p53 binds DNA as a protein homodimer but monomeric variants retain full transcription transactivation activity. Oncogene. 1993 Nov;8(11):3165–3173. [PubMed] [Google Scholar]
- Truant R., Xiao H., Ingles C. J., Greenblatt J. Direct interaction between the transcriptional activation domain of human p53 and the TATA box-binding protein. J Biol Chem. 1993 Feb 5;268(4):2284–2287. [PubMed] [Google Scholar]
- Unger T., Nau M. M., Segal S., Minna J. D. p53: a transdominant regulator of transcription whose function is ablated by mutations occurring in human cancer. EMBO J. 1992 Apr;11(4):1383–1390. doi: 10.1002/j.1460-2075.1992.tb05183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Kinzler K. W. p53 function and dysfunction. Cell. 1992 Aug 21;70(4):523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- Wang P., Reed M., Wang Y., Mayr G., Stenger J. E., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: structure, oligomerization, and transformation. Mol Cell Biol. 1994 Aug;14(8):5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Reed M., Wang P., Stenger J. E., Mayr G., Anderson M. E., Schwedes J. F., Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993 Dec;7(12B):2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Hauschka S., Tapscott S. J. The MCK enhancer contains a p53 responsive element. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):4570–4571. doi: 10.1073/pnas.88.11.4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription: in tune with the histones. Cell. 1994 Apr 8;77(1):13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- Wu X., Bayle J. H., Olson D., Levine A. J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993 Jul;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Zambetti G. P., Bargonetti J., Walker K., Prives C., Levine A. J. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992 Jul;6(7):1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Kern S. E., Pietenpol J. A., Kinzler K. W., Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992 Apr;1(1):45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993 Nov 19;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]