Fig. 4.
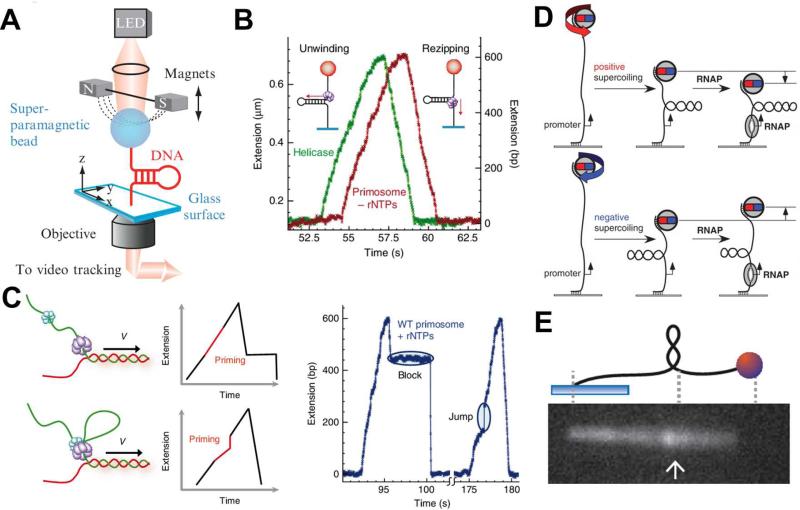
Magnetic tweezers used for single-molecule studies of DNA replication and transcription. (A) A schematic illustrating the experimental setup of magnetic tweezers. (B) Experimental traces of the activities of T4 gp41 helicase (green) and primosome (red) in the absence of rNTPs on the hairpin DNA substrate. (C) Schematic presentation of two possible models for the T4 primase and helicase interactions and the expected traces for each model (left) and two experimental traces of primosome activities on the hairpin DNA substrate in the presence of rNTPs (right). The disassembly mechanism (top): gp61 during primer synthesis dissembles with gp41, forming the primase-primer complex on DNA that “blocks” rezipping of hairpin after unwound by gp41. The priming loop mechanism (bottom): gp41 and gp61 remain associated and the DNA, when continuously unwound by gp41 during primer synthesis, forms a loop that gives a “jump” extension after it collapses. The features of “block” and “jump” in the experimental traces provide evidence for the operations of disassembly and priming loop mechanisms respectively (right). (D) Unwinding of one turn of promoter by RNA polymerase with a positively (top) or negatively (bottom) superhelical substrate give rise to a drop or an increase of the position of magnetic beads in the Z-direction. (E) Experimental setup of a hybrid system of magnetic tweezer and epifluorescence microscopy. Rotation of the bead forms the supercoiled DNA, which is subsequently pulled side-way into the focal plane of an aperture objective for fluorescence imaging. Images are reproduced from Ref. 63, 64, 69, and 70.