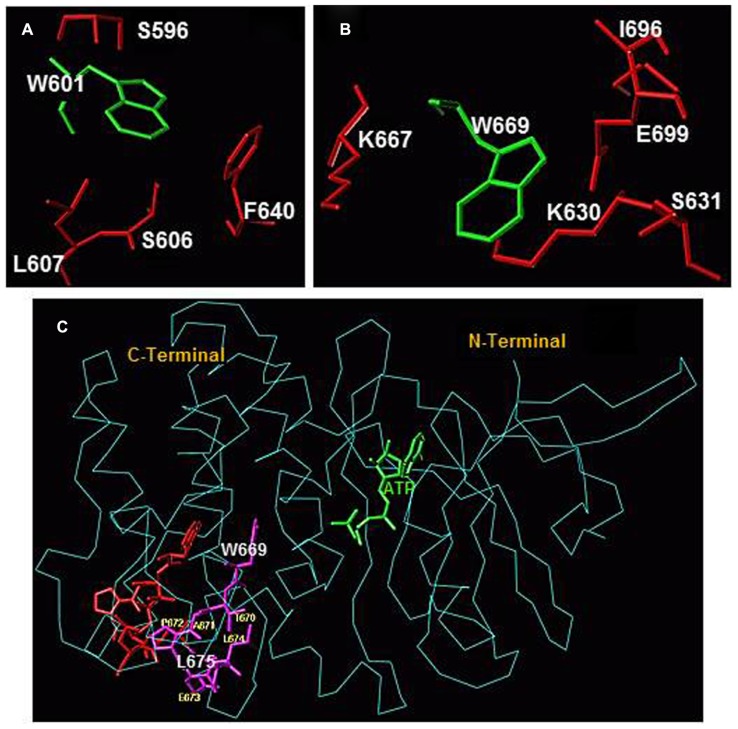
FIGURE 2.
(A) Amino acid residues surrounding W601 and (B) amino acid residues surrounding W669 within the ARM domain. Amino acid residues depicted in red are located within a 4 Å sphere from the respective tryptophan residue (green). (C) Conformational changes within the 669WTAPELL675 motif induced by ATP binding to the ARM domain. The backbone structure of the ATP-bound ARM domain is shown in cyan and the ATP molecule is in green. The 669WTAPELL675 motif is shown in magenta color. Apo structure of the ARM domain was superimposed on the ATP-bound form to assess the relative, ATP binding induced, conformational changes. For clarity, only the 669WTAPELL675 motif (shown in red) of the apo-enzyme is visible. ATP binding results in a more compact structure of the ARM domain: the W669 side chain moves toward the ATP binding pocket while the side chains of T670, E673, L674, and L675 move toward the protein surface [compare the orientation of side chain of these amino acid residues before (in red) and after (in magenta) ATP binding; fonts for W669 and L675 residues are increased for better visibility]. This movement changes the surface properties of the ARM domain. The movement toward the surface of the protein is poised to facilitate interaction of this amino acid stretch with subsequent transduction motif, possibly within the catalytic domain, propagation of the ANF/ATP binding signal and activation of the catalytic domain (reproduced with permission from ref. Duda et al., 2009).