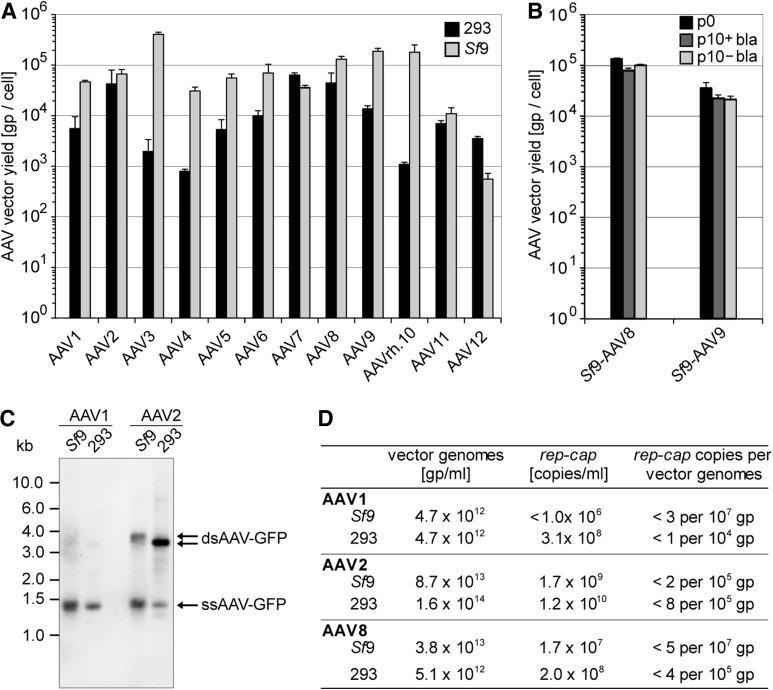
FIG. 2.
Burst sizes and integrity of AAV1–12 vectors produced in 293 cells compared with Sf9 cells. (A) The yields of rAAV1–12 preparations were quantified as benzonase-resistant genomic particles (gp) per cell by LightCycler PCR using rAAV genome-specific primers. Benzonase-treated freeze–thaw supernatants were prepared 72 hr posttransfection of 293 cells or infection of Sf9 cells, respectively. rAAV burst sizes of three experiments are displayed as mean±standard deviation. Please note the logarithmic scale of the y-axis. (B) Analysis performed similar to (A). The burst sizes of rAAVs from AAV8 and -9 Sf9 producer cell lines after serial passages with or without blasticidin selection were quantified at passage 0 (p0) and passage 10 (p10±basticidin). (C) Southern blot analysis of vector genomes from AVB sepharose-purified AAV1 and -2 vector preparations. Vector genomes are detected by a biotin-labeled probe specific for GFP. The size of the packaged rAAV genomes varies between Sf9- (pTR-UF26: 3,794 bp) and 293-cell-derived (pTR-UF5: 3,388 bp) vectors. The arrows point to ssDNA and dsDNA AAV genomes that are detected because of partial reassociation of packaged (+) and (−) strand genomes. (D) Quantitative analysis of the AAV vector genomes (vg) and copackaged wild-type rep/cap sequences of AAV1, -2, and -8.