Significance
The role of repressive gene regulation through histone modifications is known in various biological processes. Histone H3 trimethyl Lys27 (H3K27me3) represses gene expression, and our goal was to understand how this methylation regulates cell differentiation in the vertebrate retina. In this work, we focused on the role of demethylase (Jmjd3) of H3K27me3 in retinal development. Spatiotemporal expression patterns of Jmjd3 during retinal development were shown. Suppression of Jmjd3 expression during retinal development resulted in the failure of differentiation of retinal cell subsets. Lowered expression of genes essential for differentiation of the subsets by loss of expression of Jmjd3 was observed. Therefore, we propose that lineage-specific H3K27me3 demethylation of critical gene loci by spatiotemporal-specific Jmjd3 expression is required for appropriate maturation of retinal cells.
Keywords: histone methylation, progenitor, interneuron
Abstract
Di- and trimethylation of lysine 27 on histone H3 (H3K27me2/3) is an important gene repression mechanism. H3K27me2/3-specific demethylase, Jmjd3, was expressed in the inner nuclear layer during late retinal development. In contrast, H3K27 methyltransferase, Ezh2, was highly expressed in the embryonic retina but its expression decreased rapidly after birth. Jmjd3 loss of function in the developing retina resulted in failed differentiation of PKC-positive bipolar cell subsets (rod-ON-BP) and reduced transcription factor Bhlhb4 expression, which is critical for the differentiation of rod-ON-BP cells. Overexpression of Bhlhb4, but not of other BP cell-related genes, such as transcription factors Neurod and Chx10, in Jmjd3-knockdown retina rescued loss of PKC-positive BP cells. Populations of other retinal cell subsets were not significantly affected. In addition, proliferation activity and apoptotic cell number during retinal development were not affected by the loss of Jmjd3. Levels of histone H3 trimethyl Lys27 (H3K27me3) in the Bhlhb4 locus were lower in Islet-1–positive BP cells and amacrine cells than in the Islet-1–negative cell fraction. The Islet-1–negative cell fraction consisted mainly of photoreceptors, suggestive of lineage-specific demethylation of H3K27me3 in the Bhlhb4 locus. We propose that lineage-specific H3K27me3 demethylation of critical gene loci by spatiotemporal-specific Jmjd3 expression is required for appropriate maturation of retinal cells.
Methylation of basic amino acid residues in histone is an important epigenetic mechanism to regulate gene expression. Recent studies have shown that histone lysine methylation regulates gene expression by influencing the accessibility of promoter or enhancer regions to transcriptional molecules (1). Di- and trimethylation of lysine 27 on histone H3 (H3K27me2/3) by the histone methyltransferase enhancer of the zeste homolog 2 (Ezh2/Kmt6) with polycomb repressive complex 2 (PRC2) is known as a mechanism of gene repression (2–4). Histone H3 trimethyl Lys27 (H3K27me3) represses gene expression by recruiting PRC1, which recognizes H3K27me3 and ubiquitinate lysine 119 on histone H2A (5). The role of H3K27me3 markers is well established in developmental processes (6). Dynamic switches in polycomb targets that restrict pluripotency and define the developmental potential of progenitor cells have been demonstrated by differentiation of ES cells into Pax6-positive radial-glial neuronal progenitor cells (7). The role of H3K27 in defining the timing of neurogenesis and gliogenesis was shown in cortical neurons by ablating Ezh2 at different developmental stages (8–10). Jmjd3 (Kdm6b) and Utx (Kdm6a) are H3K27 demethylases (11, 12). Jmjd3 is required to regulate the maintenance of the respiratory rhythm generator during late development and in the function of neuronal networks (13).
The vertebrate neural retina is organized into a laminar structure composed of six types of neurons and glial cells. In the mouse, these major retinal cell classes are generated from retinal progenitor cells between embryonic day (E) 11 and postnatal day (P) 10 in a conserved temporal order (14). The importance of histone methylation during retinal development has been discussed in previous studies (details are provided in SI Note 1). Recently, it was reported that loss of function of ezh2 in Xenopus causes defective proliferation of retinal progenitor cells and repression of proneural gene expression (15). Xenopus ezh2 was expressed in the ganglia cell layer (GCL) and inner nuclear layer (INL) in the developing retina, and low-level expression of jmjd3 in these layers has been reported (16). However, the functional roles of the H3K27 markers in mammalian retinal development have not been reported. In the present study, we examined the roles of H3K27 markers in retinal cell differentiation by ablating Jmjd3 during retinal development. We show that timed Jmjd3 expression, which results in the demethylation of key genes and retinal maturation factors, plays a critical role in the differentiation of retinal subsets.
Results
Jmjd3 Expression in the INL of Developing Retina.
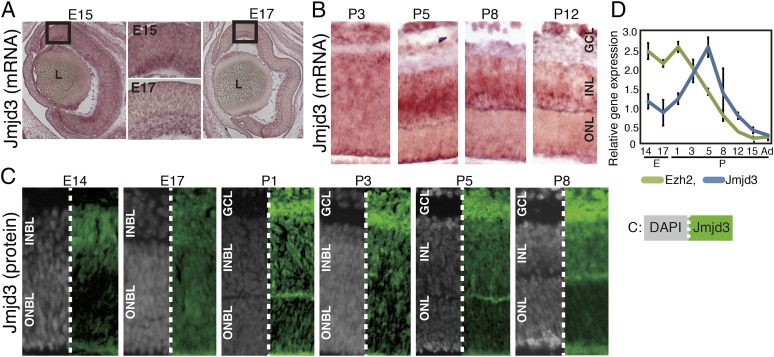
We first examined the expression patterns of enzymes related to H3K27 modification. Only one enzyme (Ezh2) is known to act as a methyltransferase, whereas three enzymes (Jmjd3, Utx, and Uty) are demethylases at the H3K27 site (17). We examined the expression patterns of Jmjd3 and Utx in the developing mouse retina using in situ hybridization (Fig. 1 A and B and Fig. S1 A and B). Jmjd3 was expressed as early as E15 in the retina, and a weak signal was observed in the inner half of the retina. At E17 and P3, the Jmjd3 signal was weak and the region of expression was difficult to identify (Fig. 1 A and B). At P5, a signal was observed in the INL and GCL. At P8 and P12, signals in the INL decreased, although a faint signal was still detected (Fig. 1B). The Utx in situ hybridization signal was very weak, and at P8 and P12, faint signals were observed in the INL (Fig. S1A, arrows). Hybridization analysis with sense probes targeting Jmjd3 gave relatively high background but did not generate specific signals, and probes for Utx gave no background or specific signals for the examined stages (Fig. S1B). We then performed immunostaining of Jmjd3 with retinal markers. At E14 and E17, Jmjd3 was expressed in GCL and at the inner side of the blastic layer (Fig. 1C). Costaining with the Ki67 proliferation antigen indicated that Jmjd3 was expressed in postmitotic cells (Fig. S1C). At P1, Jmjd3 was weakly expressed in GCL, and at P3, signals appeared at the inner neuroblastic layer (INBL) (Fig. 1C). At P5, the Müller glia marker Ccdn3 (P5) and pan-bipolar (BP) marker Chx10 (P5 and P8) colocalized with Jmjd3, but PNR (Nr2e3), which marks rod photoreceptors (18), did not in P5 and P8 (Fig. S1C). RT-quantitative PCR (qPCR) showed that Jmjd3 expression peaked at the P5 retina, and was relatively low before and after this stage (Fig. 1D). Ezh2 is known to be highly expressed in the retina in the INBL at E16 but is undetectable in adult retina (19). RT-qPCR showed that the level of Ezh2 decreased after birth, and when retinal development was complete at about 2 wk after birth, Ezh2 expression was negligible (Fig. 1D). Immunostaining showed that Ezh2 was expressed in the whole retinal area at E14 and in all layers (GCL, INL, and outer neuroblastic layer) at E17 (Fig. S1D). At P5, Ezh2 was expressed in the GCL and weakly in some INL cells. At P8, expression of Ezh2 was observed in the outer half of the INL.
Fig. 1.
Expression of Jmjd3 and Ezh2 in developing retina. In situ hybridization (A and B) and immunostaining (C) of Jmjd3 in the developing mouse retina. Mouse retinas at indicated developmental stages were frozen-sectioned, and in situ hybridization and immunostaining were done. Signals in A and B are visualized using the 5-Bromo-4-chloro-3-indolyl phosphate (BCIP) and nitro-blue tetrazolium chloride (NBT) system. In C, Jmjd3 signals are shown in green and gray signals show nuclear staining by DAPI. (D) Expression of mRNA of Jmjd3 and Ezh2 was examined by RT-qPCR using total RNA extracted from the retina at different developmental stages. An average of three independent samples is shown with the SD. Ad, adult; L, lens; ONBL, outer neuroblastic layer.
Jmjd3 Knockdown in the Developing Retina Results in Loss of Rod-ON-BP Cells.
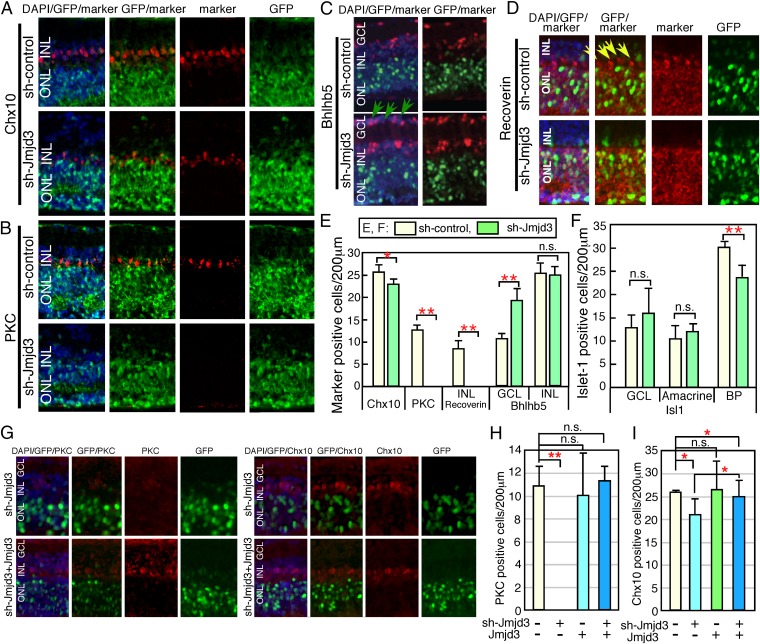
To examine the role of demethylation of H3K27 in retinal development, we performed Jmjd3 loss-of-function analysis. At E17, the retina was transfected with U6 promoter-driven shRNA for Jmjd3 (sh-Jmjd3) with an EGFP expression plasmid and cultured as explants. We first examined the effects of sh-Jmjd3 on H3K27me3 in the retina. Immunostaining (Fig. S2A) showed that the patterns of H3K27me3 in the developing retina were comparable to those in previous reports (20). Next, the effects of sh-Jmjd3 on the H3K27me3 expression pattern were examined. At day 5, signals with stronger intensity for H3K27me3, especially in the INL, were observed in sh-Jmjd3 transfected retina compared with sh-control transfected samples (Fig. S2B). At day 10, signal intensity was slightly stronger in the outer nuclear layer (ONL) (Fig. S2C). In the control, H3K27me3 signals were observed in the INL but very weakly in the ONL. However, in sh-Jmjd3–expressing retina, similar expression levels in the ONL and INL were observed (Fig. S2D). Quantification of H3K27me3-positive cells showed a higher number of positive cells in sh-Jmjd3–expressing samples than in sh-control samples at 5, 10, and 14 d of culture (Fig. S2E). We next examined retinal progenitor cell differentiation in the absence of Jmjd3 after 14 d of culture. The gross morphology of the three-layer structure of the sh-Jmjd3–expressing retina was maintained, and we examined the differentiation patterns of the retinal cell subtypes based on immunostaining. Initially, we focused on the differentiation of cells localized to the INL, because Jmjd3 was mainly expressed in the INL. Chx10 is a pan-BP marker, and the number of Chx10-positive cells in the sh-Jmjd3–expressing retina was slightly lower than in the control (Fig. 2 A and E). PKC is a marker of rod-ON-BP cells, and PKC-positive cells were almost completely absent from retinas transfected with sh-Jmjd3 (Fig. 2 B and E). Bhlhb5 is a marker of cone-OFF-BP cells and GABAergic amacrine cells, and we detected an increase in the number of displaced amacrine cells in the GCL (Fig. 2C, green arrows and Fig. 2E). In the INL, amacrine and BP cells were not clearly distinguished, and the total number of Bhlhb5-positive cells in the INL was comparable (Fig. 2E). Cone-OFF-BP cells normally express Recoverin, as seen in the control retina at the border of the ONL and INL (Fig. 2D, yellow arrows). However, Recoverin-positive Cone-OFF-BP cells were not detected in sh-Jmjd3–transfected retina (Fig. 2E). Recoverin expression in the ONL was observed in rod photoreceptors as expected (Fig. 2D). Isl1 is a marker of rod and cone-ON-BP and cholinergic amacrine cells (21, 22). The number of Isl1-positive ON-BP cells decreased by about 20% in sh-Jmjd3–transfected retina (Fig. S3A, yellow arrows and Fig. 2F). Because PKC-positive cells are a subset of Isl1-positive cells, the decrease in Isl1-positive cells likely corresponded to a decrease in PKC-positive cells. In contrast, the number of Isl1-positive amacrine cells in the INL and of displaced amacrine cells was comparable to that of the control (Fig. 2F). HuC/D is a marker of amacrine cells, and the number of displaced amacrine cells was increased in the sh-Jmjd3–transfected retina (Fig. S3B, yellow arrows), suggesting that Isl1-positive and HuC/D-positive displaced amacrine cells are at least partially different populations. We obtained similar results using a second Jmjd3 shRNA. The number of other cell types, such as Müller glia [glutamine synthetase (GS)], rod photoreceptors, and retinal ganglion cells (RGCs), were comparable to those of the control samples (Fig. S3 C–E and G). Neurite extension of RGCs was also comparable between control and sh-Jmjd3–expressing retina (Fig. S3F). We then examined the proliferation and apoptosis of retina expressing sh-Jmjd3 by examining BrdU incorporation, as well as the expression of Ki67 and active caspase-3 (Fig. S4). We transfected sh-Jmjd3 at two different retina developmental stages (E14 and E17), and we examined proliferation and apoptosis at six time points (Fig. S4A). The number of active caspase-3–positive cells at E17∼10 d showed a slightly but statistically significant difference, but no other samples showed significant differences between control and sh-Jmjd3–expressing retina (Fig. S4 B–E).
Fig. 2.
Knockdown of Jmjd3 expression during retinal development. Plasmids encoding CAG-EGFP/U6-shRNA-Jmjd3 (sh-Jmjd3)/control U6 (sh-control) (A–D), or CAG-EGFP/sh-Jmjd3/Jmjd3 expression plasmid (G–I) were introduced into the retina (E17) by electroporation. After 2 wk of explant culture, the retina was frozen-sectioned and immunostained using antibodies to antiretinal subset markers as indicated. In E, F, H, and I, the number of marker-positive cells in the electroporated region (200 μm) as judged by EGFP fluorescence was counted in sections. Green arrows in C indicate displaced amacrine cells, and yellow arrows in D indicate cone-OFF-BP cells. More than five sections from three independent samples were counted, and values with SDs are shown. **P < 0.01, *P < 0.05, and P > 0.05 (n.s.) were calculated by the Student t test. n.s., not significant.
We then examined cotransfection of Jmjd3 expression plasmids and sh-Jmjd3, and found that expression of Jmjd3 in the sh-Jmjd3–transfected retina recovered expression of PKC (Fig. 2 G and H) and Chx10 (Fig. 2 G and I). Therefore, we identified specific loss of PKC-positive rod-ON-BP cells and Recoverin-positive cone-OFF-BP cells as a consequence of Jmjd3 down-regulation.
Decreased Expression of Bhlhb4 and Vsx1 Due to Suppression of Jmjd3 Expression.
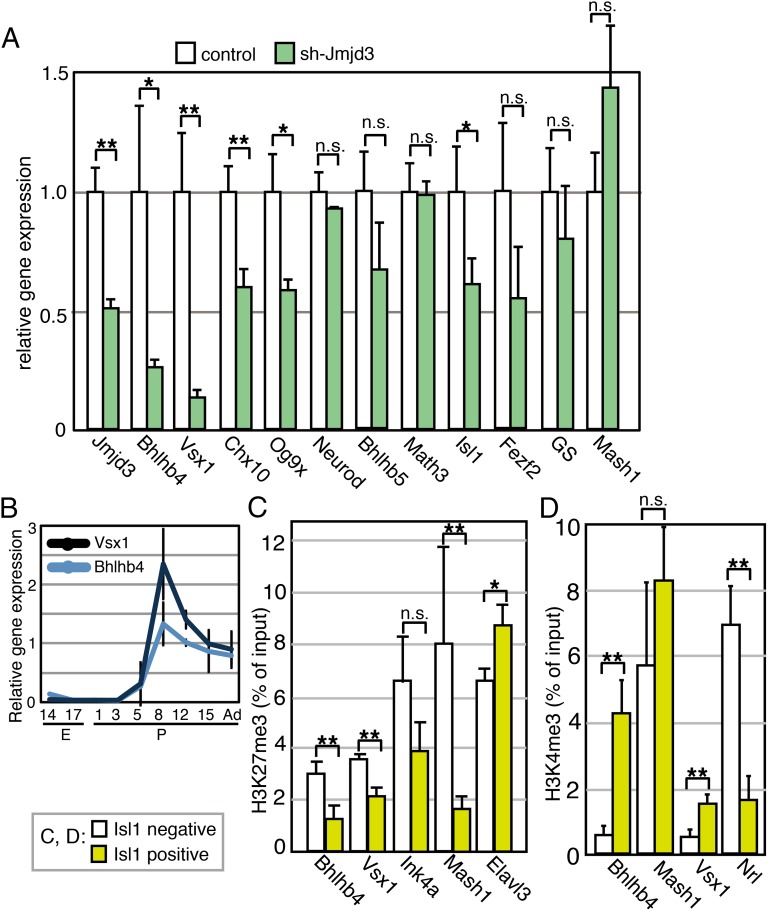
We explored whether expression of BP cell differentiation-related genes was affected by Jmjd3 down-regulation. A plasmid encoding sh-Jmjd3 was introduced into the retina at E15 and was cultured for 12 d. The expression levels of various genes were then examined by RT-qPCR (Fig. 3A). As expected, Jmjd3 expression significantly decreased. Bhlhb4 is essential for the maturation of rod-ON-BP cells, which were labeled for PKC (23), and Vsx1 plays a pivotal role in the differentiation of cone-OFF-BP cells, which were marked by Recoverin (24). We found that the expression levels of Bhlhb4 and Vsx1 were strongly suppressed in the presence of sh-Jmjd3 (Fig. 3A). Therefore, the loss of PKC-positive rod-ON-BP cells and Recoverin-positive cone-OFF-BP cells may be explained by suppressing the expression of Bhlhb4 and Vsx1, respectively, by sh-Jmjd3. The expression levels of Chx10, Og9x, and Isl1 were lower in the sh-Jmjd3–expressing cells (Fig. 3A), whereas difference of expression of the remaining genes between control and sh-Jmjd3–expressing samples was not statistically significant.
Fig. 3.
Molecular signatures in sh-Jmjd3–transfected retina. (A) Plasmids encoding sh-Jmjd3 or sh-control with CAG-EGFP were introduced into the retina at E15; the retina was then cultured for 12 d as retinal explants, and RT-qPCR was performed. All values of qPCR were normalized by the normalization factor value calculated by housekeeping genes, Gapdh and Sdha. (B) Transition of expression of Bhlhb4 and Vsx1 was examined by RT-qPCR during retinal development. H3K27me3 (C) or H3K4me3 (D) modification of gene loci in purified Isl1-positive or Isl1-negative cells was examined by ChIP analysis. Experiments were done three times independently, and the average and SD are shown. Abcam antibody (ab6002) for H3K27me3 was used in C. **P < 0.01, *P < 0.05, and P > 0.05 (n.s.) were calculated by the Student t test.
Using qPCR, we examined the transition of Bhlhb4 and Vsx1 expression during retinal development using whole retinas at various developmental stages. Bhlhb4 and Vsx1 expression was not detectable until P5, and their expression levels increased markedly with retinal development (Fig. 3B).
We next determined the levels of H3K27me3 at the Bhlhb4, Vsx1, and other BP-related gene loci in BP cells. To obtain retinal cells enriched for BP cells, we used Isl1, which is a marker of rod- and cone-ON-BP and cholinergic amacrine cells (21, 22). We stained permeabilized retinal cells at P9 with an anti-Isl1 antibody, followed by staining with a secondary antibody conjugated with phycoerythrin. The Isl1-positive and Isl1-negative cells were then purified using a cell sorter (Fig. S5 A and B), and ChIP analysis using an anti-H3K27me3 antibody was performed. The H3K27me3 level at the Bhlhb4 and Vsx1 loci was lower in Isl1-positive cells than in Isl1-negative cells (Fig. 3C). Because Vsx1 is also expressed in type 7 cone-ON-BP cells (24, 25), it is possible that differentiation of type 7 cone-ON-BP cells was defective in sh-Jmjd3–expressing retina. However, this was difficult to confirm, given the lack of a specific marker for this BP subset. Furthermore, although Isl1 is strictly expressed in the ON-BP subset in the mature retina (21, 22, 26), Isl1-KO mice also have OFF-BP subset defects, suggesting that Isl1 is transiently expressed in OFF-BP cell precursors (27) and that the precursors of Recoverin-positive cells are found in the Isl1-positive fractions. The level of H3K27me3 at the Cdkn2a (Ink4a) locus was not significantly different between the Isl1-positive and Isl1-negative populations, and that at the Elavl3 locus was slightly higher in Isl1-positive cells. The H3K27me3 level at the Mash1 locus was significantly lower in Isl1-positive cells (Fig. 3C). All other genes showed higher values in Isl1-positive cells than in Isl1-negative cells, but the difference was not statistically significant (Fig. S5C). We also examined levels of H3K27me3 at the Chx10, Neurod, and GS loci, but we could not detect reliable qPCR signals because the H3K27me3 levels at these gene loci were too low in both cell fractions.
We then explored whether the pattern of H3K27 levels in the Bhlhb4 locus is specific for H3K27me3 by examining Histone H3 trimethyl Lys4 (H3K4me3) levels. ChIP analysis of H3K4me3 using Isl1-positive/negative retinal cells at P9 was performed. Interestingly, H3K4me3 marker in the Bhlhb4 locus was much higher in the Isl1-positive fraction than in the Isl1-negative fraction (Fig. 3D), suggesting that H3K4me3 modification also contributes to specific up-regulation of Vsx1 and Bhlhb4 in cells in the Isl1-positive cell fraction. The H3K4me3 level of the Vsx1 locus was also higher in the Isl1-positive fraction, but that of Nrl, which is a photoreceptor-specific gene (28), was higher in Isl1-negative cells (Fig. 3D).
Bhlhb4 Expression Reverses the Loss of PKC-Positive Cells in sh-Jmjd3– Transfected Cells.
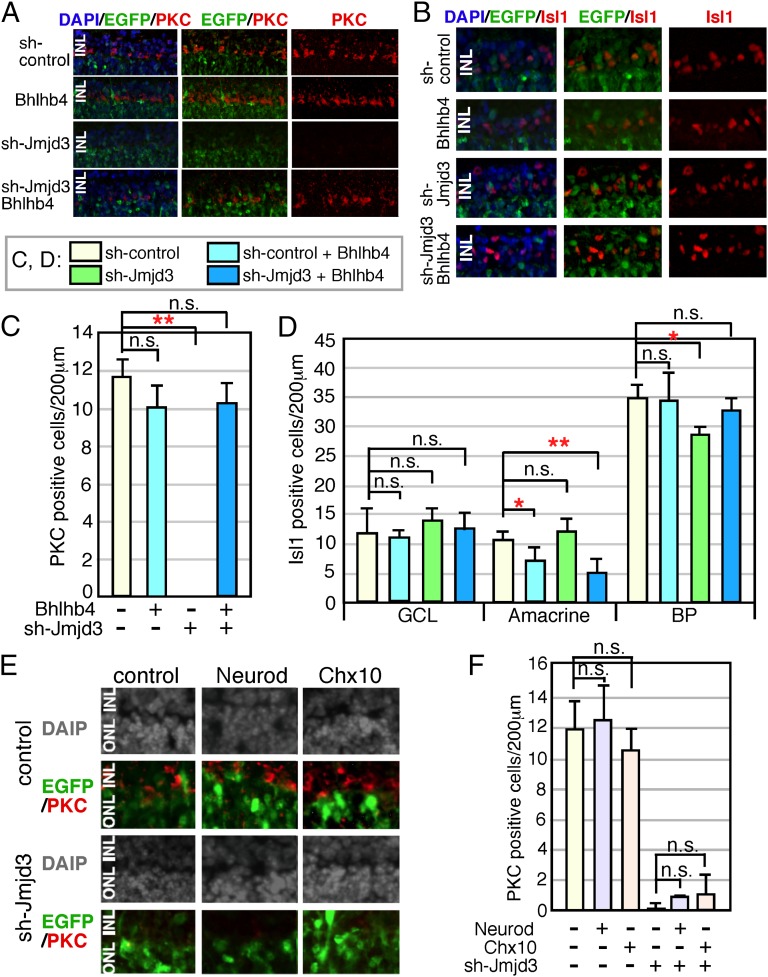
We then focused on the PKC-positive BP cells for a more detailed molecular analysis. We investigated whether Bhlhb4 caused the loss of PKC-positive cells in sh-Jmjd3–expressing retina by forcing Bhlhb4 expression in sh-Jmjd3–expressing cells. We first examined the effects of Bhlhb4 expression alone in retinal progenitor cells (Fig. 4 A–D). Bhlhb4-expressing and EGFP-expressing plasmids were transfected into the retina at E17, and the retina was cultured as an explant for 2 wk. Differentiation of the retina was then examined by immunostaining of frozen sections (Fig. 4 A and B). Forced Bhlhb4 expression did not cause apparent morphological abnormalities of the retina, and PKC immunostaining showed comparable differentiation of rod-ON-BP cells (Fig. 4 A and C). The Bhlhb4-expressing and sh-Jmjd3–expressing plasmids were then cotransfected into E17 retina. Bhlhb4 expression reversed the loss of PKC-positive cells by sh-Jmjd3 (Fig. 4 A and C), suggesting that the loss of PKC-positive cells by sh-Jmjd3 was caused by the loss of Bhlhb4 expression. The slight decrease in the number of Isl1-positive BP cells caused by sh-Jmjd3 was reversed by Bhlhb4 expression (Fig. 4 B and D). We also performed rescue experiments using Neurod and Chx10, but we did not observe PKC-positive cells by expressing Neurod or Chx10 in the presence of sh-Jmjd3 (Fig. 4 E and F).
Fig. 4.
Supplemental expression of Bhlhb4 into sh-Jmjd3–transfected retina. Mouse retina at E17 was isolated, and plasmids encoding CAG-EGFP and sh-Jmjd3 or sh-control with or without CAG-Bhlhb4 (A–D) and CAG-Neurod or CAG-Chx10 (E and F) were introduced into the retina by electroporation. The total amount of transfected plasmids was adjusted as the same for all experiments by the addition of empty vector. (A, B, and E) After 2 wk of explant culture, the retina was frozen-sectioned and immunostained using antibodies to antiretinal subset markers as indicated. (C, D, and F) Number of marker-positive cells in the electroporated region (200 μm) was counted. More than five sections from three independent samples were counted, and values with SDs are shown. **P < 0.01, *P < 0.05, and P > 0.05 (n.s.) were calculated by the Student t test.
Discussion
In the present study, we examined the role of H3K27me3 modification in retinal development and analyzed the effects of loss of function of a demethylation enzyme. We observed a critical role of Jmjd3 for the maturation of PKC-positive BP cell subset development. Because H3K27me3 suppresses gene expression (29, 30), our results were indicative of the importance of timed demethylation to activate genes responsible for proper differentiation of BP cell subsets. Down-regulation of Bhlhb4 and Vsx1 expression, both of which are critical for BP subset differentiation, occurred in the absence of Jmjd3. Thus, we propose a model in which H3K27 methylation at loci critical for BP cell maturation occurs in retinal progenitor cells, which is retained during differentiation. Timed demethylation of genes in a specific subset of cells (i.e., BP precursor cells) releases the genes from the transcriptionally silent state (Fig. S6). This is strongly supported by our findings of a lower level of H3K27me3 at critical loci in the BP-enriched cell fraction and of Jmjd3 expression in the INL of the postnatal retina. The Mash1 locus also showed a lower level of H3K27me3 in the BP-enriched cell fraction. Mash1 plays critical roles for BP cell differentiation (31, 32), therefore suggesting that this phenomenon is biologically relevant to achieve cell lineage-specific expression of Mash1. However, the mRNA level of Mash1 in sh-Jmjd3–expressing retina was comparable to that of the control, suggesting some additional mechanisms for regulation of Mash1 transcription.
In contrast to the expression pattern of Jmjd3, H3K27 methyltransferase, Ezh2 was highly expressed in the embryonic retina. These results suggest that both methylation and demethylation are based on the spatiotemporal dynamics of related enzyme expression patterns. Regulation of the expression pattern dynamics of critical genes for neural fate determination by polycomb and timed H3K27 demethylation in the CNS were suggested previously (7, 10). Furthermore, critical roles for Jmjd3 during early neural development in ES cell neurulation and chick spinal cord development have been demonstrated (33, 34). Our results suggest that during retinal differentiation, Jmjd3 directs differentiation of subsets of neurons to regulate neuron/glia cell fate. During the late phase of neural development, Jmjd3 is required in the embryonic respiratory neuronal network (13). In the immune system, Jmjd3 is induced by bacterial products and inflammatory cytokines, and it is essential for M2 macrophage polarization in response to helminth infection and chitin, which is mediated by Irf4 (35, 36). In addition, Jmjd3 is essential for proper bone marrow macrophage differentiation (36), which suggests that the important role of timed Jmjd3 expression for the maturation of specific cell subsets applies to the hematopoietic system.
We found an increased number of displaced amacrine cells (HuC/D- or Bhlhb5-positive cells) in sh-Jmjd3–treated retina. These cells are the result of failed maturation of PKC-positive BP precursor cells, although the molecular basis for this phenomenon remains unknown. The presence of other Jmjd3 targets in the retina is currently being explored. ChIP sequencing using a purified subfraction of retinal cells could be used to identify Jmjd3 targets, although refinement of the ChIP technique is required to generate reliable results from such a low number of cells. In this study, we used retinal explants in combination with sh-RNA–mediated gene suppression. This system provides an excellent model system for molecular analysis but is limited, because we cannot analyze the effects of sh-RNA on early-born retinal subsets. Because Jmjd3 was strongly expressed in the GCL, it may play a role in the development of RGCs and amacrine cells. This could be addressed by analyzing conditional KOs of Jmjd3 in the future.
Materials and Methods
All animal experiments were approved by the Animal Care Committee of the Institute of Medical Science, University of Tokyo, and conducted in accordance with the Association for Research in Vision and Ophthalmology statement for the use of animals in ophthalmic and vision research. The U6 promoter driving the shRNA vector was used for targeting Jmjd3, and the pCAG vector was used for overexpression. Plasmids were introduced into retinal explants by electroporation. Frozen sections in optimal cutting temperature compound were used for immunostaining. ChIP-qPCR and RT-qPCR results were normalized in accord with minimum information for publication of quantitative real-time PCR experiment guidelines. Further details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eli Lyons for language assistance and Honami Watanabe for technical advice. This work is supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311480111/-/DCSupplemental.
References
- 1.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 2.Hansen KH, et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10(11):1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci. 2010;35(6):323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134(2):223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 7.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30(6):755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Hirabayashi Y, et al. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63(5):600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107(36):15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Testa G. The time of timing: How Polycomb proteins regulate neurogenesis. Bioessays. 2011;33(7):519–528. doi: 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- 11.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 12.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 13.Burgold T, et al. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2012;2(5):1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Marquardt T, Gruss P. Generating neuronal diversity in the retina: One for nearly all. Trends Neurosci. 2002;25(1):32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 15.Aldiri I, Moore KB, Hutcheson DA, Zhang J, Vetter ML. Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/β-catenin signaling. Development. 2013;140(14):2867–2878. doi: 10.1242/dev.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi A, Ochi H, Sudou N, Ogino H. Comparative expression analysis of the H3K27 demethylases, JMJD3 and UTX, with the H3K27 methylase, EZH2, in Xenopus. Int J Dev Biol. 2012;56(4):295–300. doi: 10.1387/ijdb.113360ak. [DOI] [PubMed] [Google Scholar]
- 17.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131(1):29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M, et al. Identification of a photoreceptor cell-specific nuclear receptor. Proc Natl Acad Sci USA. 1999;96(9):4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao RC, et al. Dynamic patterns of histone lysine methylation in the developing retina. Invest Ophthalmol Vis Sci. 2010;51(12):6784–6792. doi: 10.1167/iovs.09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popova EY, et al. Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PLoS ONE. 2012;7(10):e46867. doi: 10.1371/journal.pone.0046867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galli-Resta L, Resta G, Tan SS, Reese BE. Mosaics of islet-1-expressing amacrine cells assembled by short-range cellular interactions. J Neurosci. 1997;17(20):7831–7838. doi: 10.1523/JNEUROSCI.17-20-07831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007;503(1):182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bramblett DE, Pennesi ME, Wu SM, Tsai MJ. The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron. 2004;43(6):779–793. doi: 10.1016/j.neuron.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 24.Chow RL, et al. Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc Natl Acad Sci USA. 2004;101(6):1754–1759. doi: 10.1073/pnas.0306520101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z, et al. Vsx1 regulates terminal differentiation of type 7 ON bipolar cells. J Neurosci. 2011;31(37):13118–13127. doi: 10.1523/JNEUROSCI.2331-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haverkamp S, Ghosh KK, Hirano AA, Wässle H. Immunocytochemical description of five bipolar cell types of the mouse retina. J Comp Neurol. 2003;455(4):463–476. doi: 10.1002/cne.10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elshatory Y, et al. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 2007;27(46):12707–12720. doi: 10.1523/JNEUROSCI.3951-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swaroop A, et al. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci USA. 1992;89(1):266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 30.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 31.Pollak J, et al. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development. 2013;140(12):2619–2631. doi: 10.1242/dev.091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gamm DM, et al. Regulation of prenatal human retinal neurosphere growth and cell fate potential by retinal pigment epithelium and Mash1. Stem Cells. 2008;26(12):3182–3193. doi: 10.1634/stemcells.2008-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akizu N, Estarás C, Guerrero L, Martí E, Martínez-Balbás MA. H3K27me3 regulates BMP activity in developing spinal cord. Development. 2010;137(17):2915–2925. doi: 10.1242/dev.049395. [DOI] [PubMed] [Google Scholar]
- 34.Burgold T, et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3(8):e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 36.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.