Significance
The development of dental plaque biofilm requires specific and sequential molecular interactions between oral bacteria-colonizing host surfaces. Coaggregation between early colonizers is crucial to establish an environment suitable for late colonizers. Here, we describe that a surface protein in a Gram-positive bacterium that is not genetically linked to the fimbrial gene clusters hijacks a specific fimbrial polymerization apparatus to be displayed on the fimbrial tip. This tip-localized protein not only functions as the bona fide cell-to-cell adhesion factor for mediating coaggregation between the early colonizers Actinomyces oris and Streptococcus oralis but also serves as an initiator of fimbrial assembly.
Keywords: sortase, pilus assembly, interbacterial interaction, cell wall-anchored proteins
Abstract
The formation of dental plaque, a highly complex biofilm that causes gingivitis and periodontitis, requires specific adherence among many oral microbes, including the coaggregation of Actinomyces oris with Streptococcus oralis that helps to seed biofilm development. Here, we report the discovery of a key coaggregation factor for this process. This protein, which we named coaggregation factor A (CafA), is one of 14 cell surface proteins with the LPXTG motif predicted in A. oris MG1, whose function was hitherto unknown. By systematic mutagenesis of each of these genes and phenotypic characterization, we found that the Actinomyces/Streptococcus coaggregation is only abolished by deletion of cafA. Subsequent biochemical and cytological experiments revealed that CafA constitutes the tip of a unique form of the type 2 fimbria long known for its role in coaggregation. The direct and predominant role of CafA in adherence is evident from the fact that CafA or an antibody against CafA inhibits coaggregation, whereas the shaft protein FimA or a polyclonal antibody against FimA has no effect. Remarkably, FimA polymerization was blocked by deletion of genes for both CafA and FimB, the previously described tip protein of the type 2 fimbria. Together, these results indicate that some surface proteins not linked to a pilus gene cluster in Gram-positive bacteria may hijack the pilus. These unique tip proteins displayed on a common pilus shaft may serve distinct physiological functions. Furthermore, the pilus shaft assembly in Gram-positive bacteria may require a tip, as is true for certain Gram-negative bacterial pili.
In Gram-positive pathogens, many virulence factors that mediate bacterial adherence, biofilm formation, and other pathogenic processes are covalently attached on the cell surface (1). Most of these virulence factors are anchored to the cell wall by a cysteine-transpeptidase enzyme called sortase, first discovered in Staphylococcus aureus (2). The cell wall anchoring mechanism of surface proteins catalyzed by sortase is conserved in Gram-positive bacteria. Substrates of sortase contain not only an N-terminal signal peptide needed for export across the cytoplasmic membrane but also a C-terminal cell wall sorting signal (CWSS) required for cell wall anchoring that is composed of an LPXTG motif, followed by a hydrophobic region and a positively charged cytoplasmic tail (3). Sortase recognizes this LPXTG motif, cleaves between the threonine and glycine residues, and joins the cleaved polypeptide to the stem peptide of the cell wall lipid II precursor that is ultimately incorporated into the cell envelope (4). In a number of Gram-positive pathogens studied to date, some LPXTG-containing proteins are assembled into covalently linked polymers known as pili (or fimbriae) by a unique class of “pilus-specific” sortases first described in Corynebacterium diphtheriae (5), and subsequently in Enterococcus faecalis, Bacillus cereus, streptococci, and Actinomyces oris among others (6–11).
A. oris is a Gram-positive pathogen that plays a pivotal role in the development of dental plaque (12). The A. oris genome encodes three sortases, two of which are organized into separate gene clusters, each containing cognate CWSS-harboring substrates that form an antigenically distinct fimbria. The fimQ-fimP-srtC1 gene cluster encodes the type 1 fimbria, which is composed of FimP polymerized into the fimbrial shaft and FimQ located at the tip (11). Similarly, the fimB-fimA-srtC2 gene cluster specifies the type 2 fimbria, which is assembled from the shaft fimbrillin FimA and the tip fimbrillin FimB (11, 13). Although all fimbrillins harbor a CWSS, the shaft fimbrillins (FimA and FimP) also contain an N-terminal pilin motif harboring a conserved lysine residue, which participates directly in the cross-linking reaction that joins each pilin subunit to another (14, 15). According to our current model, using type 2 fimbriae as an example (16), the pilus-specific sortase SrtC2 joins FimB and FimA by cross-linking the threonine residue of the FimB LPXTG motif to the lysine residue of the FimA pilin motif. Elongation of the fimbrial structure is permitted when the available FimA monomers are added to this dimeric FimB-FimA intermediate. An elongated fimbrial polymer is ultimately anchored to the cell wall, preferentially by the housekeeping sortase, SrtA, that is involved in anchoring all surface proteins to the cell wall.
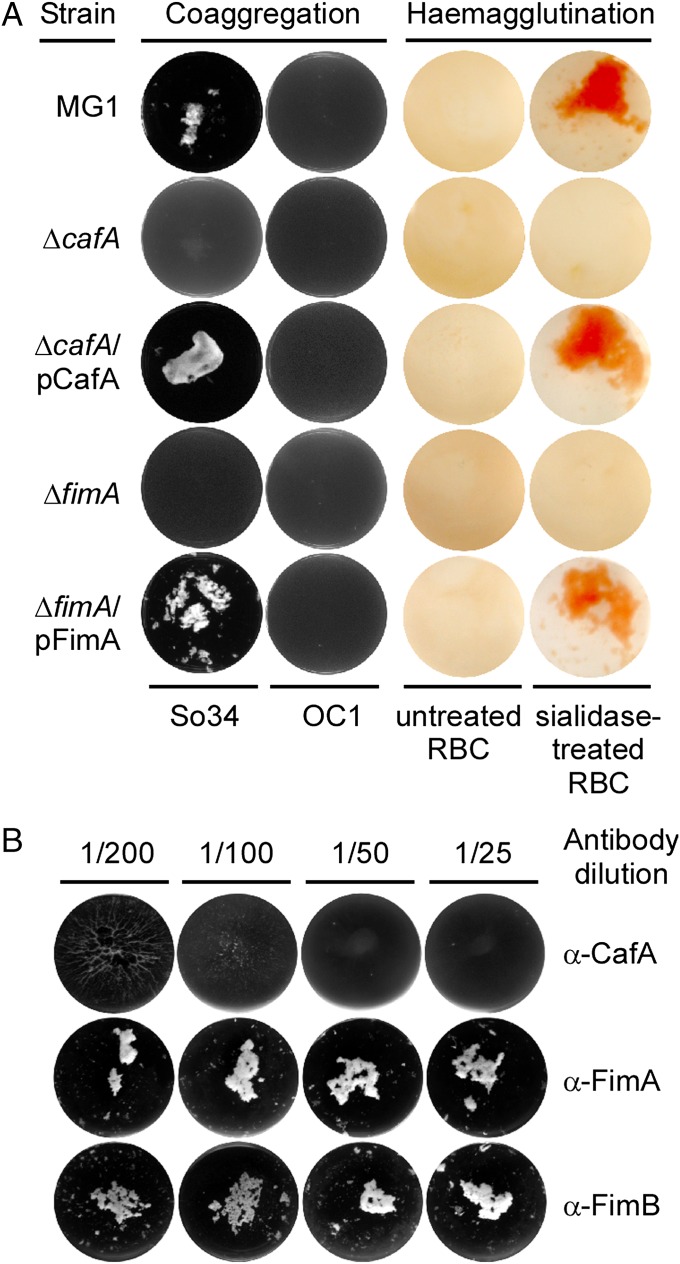
The two antigenically distinct fimbriae of A. oris perform distinct functions in pathogenesis. In the case of type 1 fimbriae, the tip fimbrillin FimQ mediates bacterial binding to the salivary proline-rich proteins that coat the tooth surface (17). Strikingly, when fimQ is deleted, the assembly of type 1 fimbriae is nearly abolished. This suggests that FimQ acts to nucleate the assembly of FimP into a fimbrial shaft. Although little is known about the determinants that govern the ordered assembly of pilins into a proper pilus structure in Gram-positive bacteria, the incorporation of a designated pilin into the pilus tip appears to rely upon the specific CWSS of the tip pilin (18). Unexpectedly, the tip fimbrillin FimB was found to be dispensable not only for assembly of the type 2 fimbrial shaft, which is made of FimA, but also for binding of the type 2 fimbriae to receptor polysaccharides (RPSs) on the surface of streptococci, resulting in the coaggregation of this bacterium with Actinomyces (13). This interbacterial interaction, which is critical for the development of the oral biofilm (19), is attributed to the shaft fimbrillin FimA because its absence abrogated bacterial coaggregation, biofilm formation, and hemaglutination (13). Indeed, structural studies revealed that FimA contains three IgG-like modules that are commonly found in Gram-positive pilins (15), and the recombinant FimA protein was able to bind to the surface of epithelial cells and S. oralis as well as to asialofetuin, a glycoprotein that contains the RPSs for Actinomyces interaction (15). Paradoxically, whereas antibodies raised against the purified type 2 fimbriae have been reported to block bacterial coaggregation (20), polyclonal antibodies directed against recombinant FimA or FimB did not display this inhibitory activity (Fig. 1). One possibility is that the type 2 fimbriae might contain an additional unknown component that serves this adhesive function. The present study was designed to test this intriguing hypothesis.
Fig. 1.
Requirement of CafA for receptor-mediated cell-to-cell interactions. (A) A. oris MG1 and its isogenic derivatives were examined for coaggregation with RPS-positive (So34) and RPS-negative (OC1) S. oralis. For hemagglutination, RBCs were treated with sialidase or mock-treated before incubation with A. oris cells. (B) A. oris cells were pretreated with increasing concentrations of antibodies raised against CafA, FimA, or FimB before examination for coaggregation.
A. oris MG1 harbors many predicted cell wall-anchored proteins that contain the LPXTG motif, as is true of many Gram-positive bacteria. To date, nothing has been described regarding the function of these putative cell surface proteins of A. oris. Here, we report the unexpected discovery that one of these predicted cell surface proteins, named coaggregation factor A (CafA), is incorporated at the tip of FimA polymers, thus resulting in a novel form of the type 2 fimbriae that do not contain the classical tip fimbrillin FimB. Importantly, we show that CafA is a bona fide and critical coaggregation factor that mediates interbacterial interactions. This provides a previously unidentified example of a virulence factor hitchhiking a fimbrial organelle for display on the bacterial surface. Remarkably, homologs of CafA are widespread, hinting that the mechanism of pilus hitchhiking for surface display of virulence factors may be common in Gram-positive bacteria.
Results
Identification of a Unique CafA of Actinomyces That Mediates Bacterial Coaggregation and Hemagglutination.
The fact that certain antibodies against type 2 fimbriae can block RPS-mediated bacterial coaggregation with S. oralis (20) but polyclonal antibodies against FimA do not (Fig. 1B) suggested that type 2 fimbriae may contain an unidentified surface antigen that mediates coaggregation. This prompted us to survey the A. oris MG1 genome systematically for additional surface proteins that may perform the adhesive function in the cell-to-cell interactions. A total of 14 ORFs, named acaA–acaN (aca for Actinomyces cell wall-anchored proteins) predicted to encode LPXTG-containing proteins not linked to the fimbrial gene clusters were identified (Table S1). Subsequently, we deleted each of these aca genes individually in the parental strain MG1 (Materials and Methods) and assessed the relative proficiency of the individual mutants for coaggregation with S. oralis 34 (So34; RPS-positive) (13). In these experiments, equivalent numbers of A. oris and S. oralis cells were mixed together, and coaggregation was visually determined. S. oralis OC1 lacking RPS was used as a control. Results showed that with the exception of the ΔacaF deletion mutant, all other mutant strains coaggregated normally with So34 (Fig. S1A), demonstrating that the product of the acaF gene is essential for coaggregation. We thus renamed acaF as cafA (caf for coaggregation factor), which is genetically distant from the type 2 fimbrial operon (Fig. S1B).
Deletion of cafA produced an RPS-dependent coaggregation defect identical to that seen with the deletion of fimA. This defect was rescued by expression of cafA on a complementing plasmid (Fig. 1A, Left). Furthermore, deletion of cafA also abolished receptor-mediated hemagglutination (Fig. 1A, Right), the property that previous studies have attributed to the shaft fimbrillin FimA of the type 2 fimbriae (13). Notably, in contrast to fimA deletion, which eliminates type 2 fimbriae from the Actinomyces cell surface and affects biofilm formation (13), deletion of cafA showed none of these defects (Fig. S1C). Hence, CafA appears to be selectively required for the coaggregation of Actinomyces with Streptococcus and hemagglutination.
Next, to examine whether CafA-mediated coaggregation involves a direct interaction between CafA and the Streptococcus receptors, we generated polyclonal antibodies against the mature CafA protein, which lacks its N-terminal signal peptide and the C-terminal CWSS. We then treated A. oris MG1 cells with various concentrations of polyclonal antibodies against CafA, FimA, or FimB before subjecting them to the coaggregation assay. As shown in Fig. 1B, CafA antibodies blocked A. oris coaggregation with S. oralis in a concentration-dependent manner, whereas the antibodies against FimA or FimB failed to show any measurable inhibition in this assay. Conversely, incubation of S. oralis cells with increasing amounts of the CafA protein, not the FimA protein, also prevented streptococcal coaggregation with A. oris MG1 (Fig. S2). Of note, coaggregation was not observed between S. oralis and A. oris lacking cafA (i.e., ΔcafA) when the CafA protein was added exogenously (Fig. S2). We thus conclude that CafA is the specific and predominant adherence factor that is required for A. oris coaggregation with S. oralis.
CafA Is a Component of the Type 2 Fimbrial Structures Assembled on the Bacterial Surface.
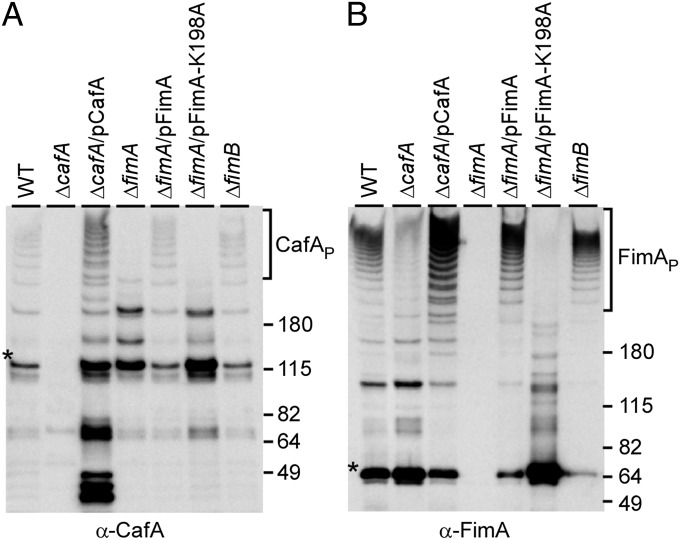
Based on above results, one might predict that CafA is a simple cell wall-associated adhesin that mediates the interbacterial interaction between Actinomyces and Streptococcus. However, the fact that the cell aggregation phenotype of the ΔcafA mutant mirrors that of the ΔfimA mutant made us wonder whether CafA is a component of the type 2 fimbriae. To examine this unprecedented scenario, we used the polyclonal anti-CafA antibody to detect surface expression of CafA through Western blotting and immunoelectron microscopy (IEM). In our quantitative Western blotting experiments, cell wall extracts of Actinomyces isolated by muramidase treatment were precipitated by trichloroacetic acid and dissolved in SDS-containing sample buffer. Protein samples representing equivalent amounts of the cell cultures were then separated on gradient gels and immunoblotted with specific antibodies (α-CafA, α-FimA, or α-FimP). Excitingly, we observed high-molecular weight polymers of CafA (CafAP) in the MG1 strain, reminiscent of the heterogeneous lengths of fimbrial polymers detected by α-FimA (compare Fig. 2 A and B, lane WT). In addition to the CafAP, monomers (predicted molecular mass of 100 kDa) and possibly dimeric forms of CafA could be detected migrating around 115 and 200 kDa, respectively (Fig. 2A). These various forms of CafA were specific to the protein because they were not observed in the ΔcafA mutant and they were restored in the complementing strain (Fig. 2A, lanes ΔcafA and ΔcafA/pCafA). Significantly, formation of the high-molecular weight CafA polymers depended on the ability of bacteria to assemble FimA polymers (Fig. 2 A and B, lanes ΔfimA and ΔfimA/pFimA). Consistent with this, CafA polymers were not detected in a lysine-substituted FimA mutant (15) that cannot polymerize the type 2 fimbriae due to its inability to form the cross-linking isopeptide bond (Fig. 2 A and B, lane ΔfimA/pFimA-K198A). Importantly, no reduction in CafA polymers was observed in a strain lacking fimB, which encodes the canonical type 2 tip fimbrillin FimB (Fig. 2 A and B, lane ΔfimB). Finally, there was a significant reduction of FimA polymers when cafA was deleted (Fig. 2B, lane ΔcafA). The observed phenotypes were specific for the type 2 fimbriae, with no effect on the type 1 fimbriae (Fig. S3). Consistently, deletion of the genes that encode components and machinery for the type 1 fimbriae did not affect the polymerization of FimA and assembly of type 2 fimbriae (11). We conclude that CafA is a covalently linked component of the type 2 fimbriae.
Fig. 2.
Polymerization of CafA and its association with FimA polymers. W fractions of A. oris MG1 (WT) and its isogenic derivatives grown to midlog phase were isolated by muramidase treatment. Equivalent protein samples were separated on 3–12% Tris⋅glycine gradient gels and detected by immunoblotting with α-CafA (A) or α-FimA (B). Protein monomers (*), polymers (P), and molecular mass markers (kilodaltons) are indicated.
CafA Forms a Distinct Fimbrial Tip Independent of FimB.
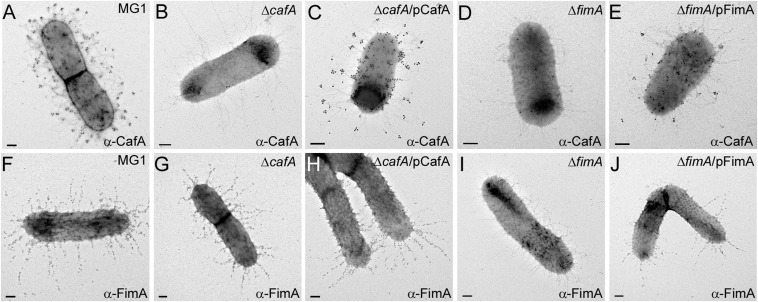
To examine how CafA is incorporated into the type 2 fimbriae, we analyzed fimbrial assembly by IEM accordingly (15). In these experiments, Actinomyces cells were incubated with specific antibodies (α-CafA and α-FimA), followed by labeling of antibody-bound cells with IgG-conjugated gold particles, and were viewed with a transmission electron microscope. Reminiscent of a typical picture obtained for labeling FimB-containing fimbriae (11, 13), the gold particles labeling CafA were detected at the outer ends of fibers extending from the bacterial envelope in MG1 cells, whereas such signals were absent on mutant bacteria in which either cafA or fimA was deleted (Fig. 3 A, B, and D); note that the fibers visible in the ΔfimA mutant are known to be type 1 fimbriae (11, 13). The missing CafA-specific gold label in strains ΔcafA and ΔfimA was restored when each protein in the corresponding mutants was expressed by trans-complementation using respective recombinant plasmids (Fig. 3 C and E).
Fig. 3.
Localization of CafA on the type 2 fimbrial structures. Cells were immobilized on nickel-carbon grids and stained with antibodies against CafA (A–E) or FimA (F–J), followed by goat anti-rabbit IgG conjugated to 12-nm gold particles. Grids were stained with 1% uranyl-acetate and viewed with an electron microscope. (Scale bars: 0.2 μm.)
To obtain further evidence that CafA is located at the tip of the type 2 fimbriae, we used double-labeling IEM, whereby fimbrial components were differentially labeled with different sizes of gold particles (11). In the parental MG1 cells, CafA stained with 18-nm gold particles was seen at the tip of FimA structures stained with 12-nm gold particles (Fig. S4A, black arrowheads). In the absence of cafA, only FimA-labeled structures were detected (Fig. S4D). Consistent with our previous report of FimB fimbrial tip localization (11), FimB labeled with 18-nm gold particles was seen at the tip of FimA structures regardless of whether CafA was present or not (Fig. S4 B and E, white arrowheads), suggesting that FimB and CafA are not colocalized on the same fimbrial structures. Consistent with this conclusion, the 18-nm gold particles labeling CafA were mostly well separated from 12-nm gold particles that specifically labeled FimB (Fig. S4 C and F).
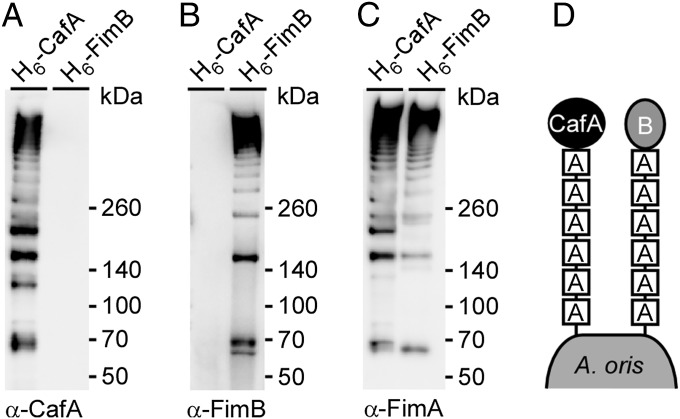
To address unequivocally whether CafA and FimB are cross-linked to separate FimA polymers, we engineered recombinant CafA and FimB proteins, with each having a His tag inserted upstream of the LPXTG motif for pull-down assays. Each construct was introduced into a corresponding deletion mutant. The cell wall extracts of these strains were isolated by muramidase treatment, and CafA and FimB proteins were purified by affinity chromatography. The eluates collected were then blotted with specific antibodies (i.e., α-CafA, α-FimA, α-FimB) to determine the nature of the purified proteins (Fig. 4). As expected, purified CafA polymers were positive for CafA- and FimA-reactive signals; importantly, these polymers did not contain FimB. Conversely, purified FimB polymers contained FimA but not CafA. Collectively, these results demonstrate that the fimbrial shaft FimA forms two distinct heterodimeric fimbrial structures, one harboring FimB and the other CafA as tip fimbrillins.
Fig. 4.
Distinct fimbrial polymers formed by CafA and FimA independent of FimB and FimA polymers. Cell wall extracts of A. oris strain ΔcafA or ΔfimB expressing CafA or FimB, respectively, with a “6×-His tag inserted upstream of the LPXTG motif, were used for affinity chromatography. Purified proteins were subjected to immunoblotting with α-CafA (A), α-FimB (B), or α-FimA (C). (D) Schematic representation shows that A. oris assembles two distinct fimbrial structures made of FimA, forming the fimbrial shaft, and CafA or FimB, each constituting the fimbrial tip.
Structural Determinants of CafA Required for Its Fimbrial Assembly and Coaggregation.
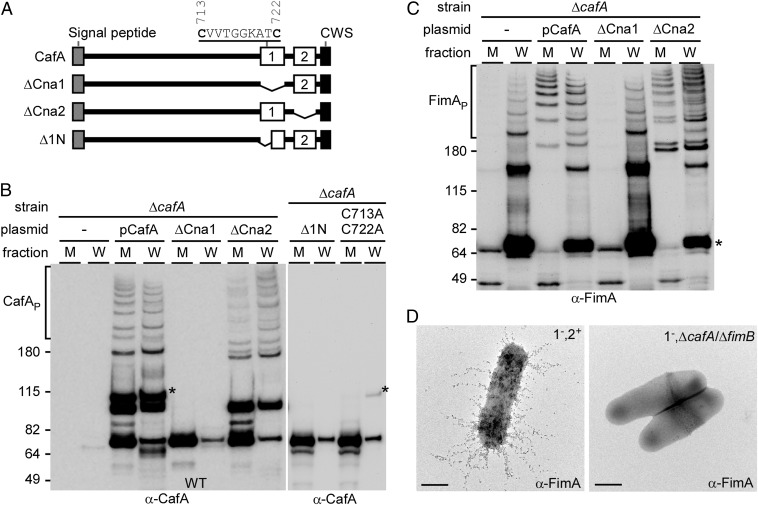
CafA is predicted to harbor two CnaB-like domains at the C terminus, named Cna1 and Cna2 (Fig. 5A). First identified in S. aureus, CnaB domains have been suggested to serve as “stalks” to orient receptor-binding regions of proteins away from the cell surface (21). To assess whether CnaB folds are important for CafA surface display and/or function, truncations of the two CnaB domains were generated and the resulting constructs were examined by immunoblot analysis and coaggregation assays. For fimbrial polymerization analysis, protein samples collected from the culture medium (M) and cell wall (W) fractions of various A. oris strains were subjected to immunoblotting with α-CafA and α-FimA as mentioned previously. Compared with the strain that expressed WT CafA, which produced CafA polymers and monomers found in the M and W fractions, a mutant carrying a version of CafA with a deletion of Cna1 failed to assemble CafA polymers but, instead, secreted CafA into the culture medium in the form of degradation products (Fig. 5B; compare lanes pCafA and ΔCna1). In contrast, deletion of Cna2 did not affect CafA incorporation into the fimbriae (Fig. 5B, lanes ΔCna2). A close inspection of the protein sequence revealed that the Cna1 domain contained a pair of cysteine residues (Fig. 5A). Mutations of these residues to alanine also caused secretion of CafA degradation products into the culture medium, a phenotype that is comparable to the deletion of Cna1 (Fig. 5B, lanes Δ1N and C713A/C722A). Consistently, the pilus polymerization defects by mutations of the Cna1 domain and cysteine residues C713/C722 paralleled the coaggregation defect of these A. oris mutants with S. oralis (Fig. S2).
Fig. 5.
Structural determinants of CafA required for fimbrial assembly and cell-to-cell interaction. (A) Diagram of CafA with a signal peptide, two CnaB-type domains with domain 1 containing a pair of cysteine residues, and a CWSS (CWS). Truncated derivatives of CafA lacking individual CnaB-type domains are shown. Protein samples collected from M and W fractions of A. oris strains carrying specific CafA mutations were analyzed by immunoblotting with α-CafA (B) or α-FimA (C). Polymers (P), monomers (*), and molecular mass markers (kilodaltons) are indicated. (D) Bacterial strains were immobilized on nickel-coated carbon grids and stained with α-FimA, followed by goat anti-rabbit IgG conjugated to 12-nm gold particles. (Scale bars: 0.2 μm.)
Interestingly, we noted that pilus polymerization in strains ΔcafA and ΔCna1 was different from that in strains pCafA and ΔCna2. In the former, no pilus polymers were found in the culture medium (Fig. 5C), suggesting that the tip fimbrillin may affect the process of pilus polymerization, similar to the phenotype of the tip fimbrillin FimQ mutant affecting assembly of the type 1 fimbriae (17). Given the fact that FimA forms two independent fimbrial structures, each with a distinct tip fimbrillin (i.e., FimB, CafA) (Fig. 4), we hypothesized that the presence of either tip fimbrillin would compensate for the loss of the other in fimbrial assembly. To investigate this, we examined pilus assembly of FimA by IEM using individual deletion mutants of cafA and fimB as well as a double mutant of cafA and fimB in the type 1 fimbria-negative background (denoted as 1−). Remarkably, although the number of FimA-labeled fimbriae appeared to be reduced in the absence of cafA or fimB (Fig. S5A, compare panel 1−,2+ with panels 1−,ΔcafA and 1−,ΔfimB), no FimA-labeled fimbriae were detected in the absence of both CafA and FimB (Fig. 5D, panel 1−,ΔcafA/ΔfimB). This was confirmed by the lack of FimA polymerization in the ΔcafA/ΔfimB mutant as detected by Western blotting (Fig. S5). Evidently, the necessity of the tip fimbrillins in fimbrial assembly is a general feature in Actinomyces, whereas the tip pilins are dispensable for the assembly of the three antigenically distinct types of pili in C. diphtheriae (5, 22, 23).
Discussion
In this work, we report the discovery of a bacterial coaggregation factor that hijacks the tip of a pilus fiber for specific interactions between two pioneer bacteria that seed the development of dental plaque, the most complex biofilm known to date. This factor, termed CafA, is one of the 14 predicted cell surface proteins of A. oris (strain MG1) not linked to the two fimbrial gene clusters and whose function had not been assessed. To address the function of these predicted proteins, we deleted each of the respective protein-coding genes individually and then investigated whether any one of the mutants affected the known coaggregation process between A. oris and S. oralis. Only one deletion mutant, ΔcafA, showed a coaggregation defect, leading to the discovery of a previously unidentified adhesin specifically involved in a key step in the initiation of oral biofilm development. Significantly, as determined by BLAST analysis, CafA is a highly conserved protein found in many Actinomyces species. Our findings thus have implications for potential therapeutic intervention.
Because constant sheer forces pose a physical challenge for bacteria colonizing the oral cavity, a tight cell-to-cell interaction or coaggregation between various colonizing bacteria may not only present a metabolic advantage over planktonic cells but directly facilitate the development of oral biofilms (24). Over 30 y ago, Cisar and coworkers (20, 25) identified the type 2 fimbriae as essential determinants for Actinomyces coaggregation to oral streptococci. This was based on the observations that an Actinomyces mutant lacking type 2 fimbriae failed to coaggregate with S. oralis (25) and that this cell-to-cell interaction was blocked by certain antibodies raised against the type 2 fimbriae (20). More recently, following the sequencing of the Actinomyces MG1 genome, genetic and biochemical work done in our laboratory revealed that the type 2 fimbriae are composed of a shaft protein FimA and a tip fimbrillin FimB that are genetically linked together in a fimbrial gene cluster encoding the fimbria-specific sortase (11). Gene deletion experiments led us to conclude that the receptor-mediated coaggregation may require the shaft protein FimA but not the tip protein FimB (13, 15). Although our in vitro experiments suggested a direct interaction between recombinant FimA of Actinomyces with surface receptors of oral Streptococcus (15), it was rather unusual that the polyclonal antibodies we raised against FimA (or another antiserum that was generated by the Cisar laboratory against the type 2 fimbria (20), which cross-reacts with FimA) do not prevent this receptor-mediated coaggregation process (Fig. 1B). The current work has essentially solved this paradox: The key adhesin that allows coaggregation is not FimA, but CafA, whose antibody does prevent bacterial coaggregation. Most importantly, we showed that CafA forms a unique tip of the type 2 fimbriae. Thus, the type 2 fimbriae are assembled in two distinct forms: one that contains FimB, whose function remains to be investigated, and the other harboring CafA, which is indeed the pilus that takes part in the coaggregation process.
Two pieces of evidence lend further support to the surprising conclusion that CafA is the actual coaggregation factor for A. oris. First, when cafA is deleted from the bacterial chromosome, the A. oris mutant cells fail to adhere to either S. oralis or RBCs, which are known to share a common RPS with that of S. oralis (Fig. 1A). Second, the coaggregation is not observed when the receptors are absent from the cell surface of S. oralis and RBCs (Fig. 1A) and CafA antibody or soluble CafA blocks the binding of bacterial surface-linked CafA to these receptors (Fig. 1B and Fig. S2A). Thus, CafA directly and specifically interacts with the cell receptors. Interestingly, CafA function is essential for bacterial coaggregation but not for biofilm formation in the presence of sucrose (Fig. S1). This CafA-independent but sucrose-dependent biofilm development is attributed to FimA, which explains the independent ability of FimA to bind polysaccharides in vitro (15), as well as the resistance of coaggregation to the polyclonal antibody raised against FimA.
An unprecedented observation reported in this paper is that CafA is associated with FimA structures, forming a distinct tip independent of the canonical tip FimB, whose gene is linked to the fimbrial gene cluster. This raises a significant question: What makes CafA unique among all other cell surface proteins to become a component of the type 2 fimbriae? Of the 14 putative surface proteins with the CWSS encoded by the A. oris MG1 genome (Table S1), it is only CafA that shows the highest homology to FimB (Expect value of 5 × 10−79 and 36% identities, based on their primary sequences). We hypothesize that CafA may fold into a tertiary structure similar to that of FimB to provide the essential determinants recognized by the pilus-specific sortase SrtC2 for fimbrial tip localization. Although this hypothesis remains to be tested experimentally, it is noteworthy that the C-terminal CWSSs of FimB and CafA are highly similar to each other, specially the presence of the FLIAGxxV motif that is absent from other Aca proteins (Fig. S1B and Table S1). This is consistent with our proposal that the CWSS is a major determinant of pilins to serve as the tip that nucleates the assembly of the pilus shaft (14). Interestingly, in this paper, we have also provided compelling evidence that the tip serves as an essential component to initiate fimbrial shaft assembly in Actinomyces: The deletion of both cafA and fimB results in the absence of FimA polymers on the cell surface (Fig. 5D). Whether or not this reflects a general rule for pilus assembly in Gram-positive bacteria is an important question that remains to be addressed in future. However, the essentiality of a tip fimbrillin in fimbrial assembly has also been observed in the case of the type 1 fimbriae of A. oris (17) and pili of Streptococcus suis (26), suggesting a conserved mechanism for fimbrial polymerization in these organisms. It is important to point out that nothing is known at present about how the pilus tip dictates and nucleates the assembly of the shaft or what governs the order in which a pilus polymer is assembled from the various monomeric pilin precursors. Equally puzzling is how expression of FimB or CafA affects fimbrial assembly or whether the expression of either is subject to regulation. Although there is no apparent regulatory element associated with the fimB gene cluster, several genes encoding AraC-type transcriptional regulators are linked to cafA and transcribed in the opposite direction from cafA. It remains to be investigated whether these regulators are genetically linked to cafA and control CafA expression and, hence, fimbrial incorporation.
Lastly, an intriguing question emerging from this work is why might there be a reason to display CafA on the bacterial surface via long fimbrial structures. We found that CafA can be anchored as monomers to the cell wall when FimA is present or absent (Figs. 2 and 3), yet the CafA monomeric molecules fail to mediate bacterial coaggregation (Fig. 1A). The simplest interpretation of this finding is that Actinomyces must have evolved pilus hijacking as a strategy to lengthen the reach of the coaggregation factors to ensure an efficient contact with the cognate receptors on the surface of the cocolonizers of the oral cavity. In the absence of this long-distance interaction, the two interacting partners will have to come into close contact with each other, which must be much less efficient stochastically than the long-distance contacts.
Materials and Methods
Strains, plasmids, and primers used in this study are listed in Tables S2–S4 of the Supporting Information, which contains information on recombinant plasmids, gene deletion, protein purification, cell fractionation and Western blotting, IEM, biofilm formation, and coaggregation assays. A. oris was grown in heart infusion (HI) broth or on HI agar plates. S. oralis was grown in HI broth supplemented with 1% glucose, whereas Escherichia coli cells were cultivated in Luria broth. When required, kanamycin was added at a concentration of 50 μg/mL.
Supplementary Material
Acknowledgments
We thank our laboratory members for critical review of the manuscript and discussion. M.E.R.-R. was supported by the Predoctoral Training Program in Molecular Basis of Infectious Diseases, National Institute of Allergy and Infectious Diseases [National Institutes of Health (NIH) Grant T32 AI55449]. This work was supported by the National Institute of Dental and Craniofacial Research of the NIH under Award F31DE024004 (to M.E.R.-R.) and Award DE017382 (to H.T.-T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321417111/-/DCSupplemental.
References
- 1.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63(1):174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285(5428):760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 3.Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40(5):1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 4.Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694(1-3):269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50(4):1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 6.Nallapareddy SR, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzik JM, Marraffini LA, Schneewind O. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol Microbiol. 2007;66(2):495–510. doi: 10.1111/j.1365-2958.2007.05939.x. [DOI] [PubMed] [Google Scholar]
- 8.Dramsi S, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60(6):1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 9.Barocchi MA, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103(8):2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauer P, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309(5731):105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 11.Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189(8):3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung MK. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10(2):120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 13.Mishra A, et al. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010;77(4):841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53(1):251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 15.Mishra A, et al. Two autonomous structural modules in the fimbrial shaft adhesin FimA mediate Actinomyces interactions with streptococci and host cells during oral biofilm development. Mol Microbiol. 2011;81(5):1205–1220. doi: 10.1111/j.1365-2958.2011.07745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ton-That H, Das A, Mishra A. Actinomyces oris Fimbriae: an Adhesive Principle in Bacterial Biofilms and Tissue Tropism. In: Kolenbrander PE, editor. Oral Microbial Communities: Genomic Inquiry and Interspecies Communication. Washington, DC: ASM Press; 2011. pp. 63–77. [Google Scholar]
- 17.Wu C, et al. Dual function of a tip fimbrillin of Actinomyces in fimbrial assembly and receptor binding. J Bacteriol. 2011;193(13):3197–3206. doi: 10.1128/JB.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quigley BR, et al. A foreign protein incorporated on the Tip of T3 pili in Lactococcus lactis elicits systemic and mucosal immunity. Infect Immun. 2010;78(3):1294–1303. doi: 10.1128/IAI.01037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 20.Revis GJ, Vatter AE, Crowle AJ, Cisar JO. Antibodies against the Ag2 fimbriae of Actinomyces viscosus T14V inhibit lactose-sensitive bacterial adherence. Infect Immun. 1982;36(3):1217–1222. doi: 10.1128/iai.36.3.1217-1222.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krishnan V, Narayana SV. Crystallography of gram-positive bacterial adhesins. Adv Exp Med Biol. 2011;715:175–195. doi: 10.1007/978-94-007-0940-9_11. [DOI] [PubMed] [Google Scholar]
- 22.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188(4):1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188(17):6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rickard AH, et al. Autoinducer 2: A concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60(6):1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- 25.Cisar JO, Curl SH, Kolenbrander PE, Vatter AE. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosus T14V. Infect Immun. 1983;40(2):759–765. doi: 10.1128/iai.40.2.759-765.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okura M, et al. The minor pilin subunit Sgp2 is necessary for assembly of the pilus encoded by the srtG cluster of Streptococcus suis. J Bacteriol. 2011;193(4):822–831. doi: 10.1128/JB.01555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.