Abstract
Objectives
Methicillin resistance in Staphylococcus spp. results from the expression of an alternative penicillin-binding protein 2a (encoded by mecA) with a low affinity for β-lactam antibiotics. Recently, a novel variant of mecA known as mecC (formerly mecALGA251) was identified in Staphylococcus aureus isolates from both humans and animals. In this study, we identified two Staphylococcus sciuri subsp. carnaticus isolates from bovine infections that harbour three different mecA homologues: mecA, mecA1 and mecC.
Methods
We subjected the two isolates to whole-genome sequencing to further understand the genetic context of the mec-containing region. We also used PCR and RT–PCR to investigate the excision and expression of the SCCmec element and mec genes, respectively.
Results
Whole-genome sequencing revealed a novel hybrid SCCmec region at the orfX locus consisting of a class E mec complex (mecI-mecR1-mecC1-blaZ) located immediately downstream of a staphylococcal cassette chromosome mec (SCCmec) type VII element. A second SCCmec attL site (attL2), which was imperfect, was present downstream of the mecC region. PCR analysis of stationary-phase cultures showed that both the SCCmec type VII element and a hybrid SCCmec-mecC element were capable of excision from the genome and forming a circular intermediate. Transcriptional analysis showed that mecC and mecA, but not mecA1, were both expressed in liquid culture supplemented with oxacillin.
Conclusions
Overall, this study further highlights that a range of staphylococcal species harbour the mecC gene and furthers the view that coagulase-negative staphylococci associated with animals may act as reservoirs of antibiotic resistance genes for more pathogenic staphylococcal species.
Keywords: β-lactams, MRSA, mecA
Introduction
A wide range of staphylococcal species harbour the mecA gene encoding an alternative penicillin-binding protein 2a (PBP2a), which has a low affinity for β-lactam antibiotics and allows cell wall synthesis to occur in the presence of β-lactam antibiotics.1–4 mecA, along with its cognate regulators mecI-mecR1, are acquired as part of a larger mobile element known as staphylococcal cassette chromosome mec (SCCmec).5 SCCmec elements insert into the chromosome at the 3′ end of the orfX by site-specific recombination mediated by the CcrA and CcrB recombinases encoded on SCCmec.6,7 Coagulase-negative staphylococcal species are thought to be the source of mecA for methicillin-resistant Staphylococcus aureus (MRSA), with a number of studies having identified likely in vivo transfer events from a coagulase-negative staphylococcal species to S. aureus.8–10 The evolutionary origins of the mecA gene are thought to lie in the common ancestor of Staphylococcus fleurettii, Staphylococcus vitulinus and Staphylococcus sciuri,11–13 further supported by experimental evidence that the mecA1 (pbpD) gene of S. sciuri is capable of mediating high-level β-lactam resistance in S. aureus.13
Recently, a novel allele of mecA was identified in MRSA from both humans and a range of animal species (livestock, small mammals and birds) across Europe.14–19 Further work in Denmark identified likely transmission events between livestock and humans, suggesting a zoonotic reservoir for the human isolates.20,21 This type of mec is named mecC (originally mecALGA251) and shares 70% nucleotide identity with mecA.18,22 The mecC gene is present with its cognate regulators mecI-mecR1 as part of a class E mec complex that shares structural similarity (mecI-mecR1-mecC-blaZ) with a mec gene complex found in Macrococcus caseolyticus.23 The class E complex is present as part of a larger 29.4 kb SCCmec type XI inserted at orfX, which also encodes the recombinase genes ccrA/B and arsenic resistance genes.18 We recently described an isolate of Staphylococcus xylosus with a novel allotype of mecC (mecC1) present as part of a possible ancestral SCCmec element.24 In this work, we describe two S. sciuri subsp. carnaticus isolates cultured from skin infection in cattle that harbour three distinct types of the mec gene (mecC, mecA and mecA1). This is the first demonstration of mecC in S. sciuri and suggests that, like the ‘conventional’ mecA gene, mecC is also present in a range of staphylococcal species found in animals. This isolate also carries a novel hybrid SCCmec consisting of SCCmec type VII, encoding mecA and a separate mecC region.
Materials and methods
Bacterial strains and growth conditions
Isolates were grown on blood agar (Oxoid, UK) and in tryptone soya broth (TSB) at 37°C. A list of isolates used in this study is shown in Table 1. Antimicrobial susceptibility testing was performed using disc susceptibility testing according to BSAC criteria (BSAC Methods for Antimicrobial Susceptibility Testing Version 11.1 May 2012). Isolates were tested for resistance to oxacillin, chloramphenicol, erythromycin, cefoxitin, ciprofloxacin, penicillin, neomycin, tetracycline, fusidic acid and gentamicin. NCTC 12493 and NCTC 6571 were used, respectively, as control resistant and susceptible isolates for oxacillin and cefoxitin.
Table 1.
Isolates of S. sciuri subsp. carnaticus and key genotypic and phenotypic characteristics described in this study
| Isolate | Resistance genotypea | Resistance phenotypeb | Reference |
|---|---|---|---|
| GVGS2 | str, blaZ, mecA, mecC, mecA1, erm(C), fexA, tet(K) | OXA, CEF, CHL, PEN, TET, FUS | this work |
| GVGS3 | str, blaZ, mecA, mecC, mecA1, fexA, tet(K) | OXA, CEF, CHL, PEN, TET, FUS | this work |
astr, streptomycin resistance; blaZ, β-lactamase (penicillin resistance); mecA, β-lactam resistance; mecC, β-lactam resistance; mecA1, potential for β-lactam resistance with a promoter mutation;51 fexA, chloramphenicol resistance; tet(K), tetracycline resistance; erm(C), erythromycin resistance.
bOXA, oxacillin; CEF, cefoxitin; CHL, chloramphenicol; PEN, penicillin; TET, tetracycline; FUS, fusidic acid.
Whole-genome sequencing
Genomic DNA of S. sciuri isolates GVGS2 and GVGS3 was extracted from overnight cultures grown in TSB at 37°C using the MasterPure Gram Positive DNA Purification Kit (Cambio, UK) or by the isothiocyanate/guanidine method.25 Illumina library preparation was carried out as described by Quail et al.26 and Hi-Seq sequencing was carried out following the manufacturer's standard protocols (Illumina, Inc., USA).
Sequence analysis and phylogenetics
Contigs for GVGS2 were assembled de novo from Fastqs with Velvet.27 Contigs containing the orfX region were closed by PCR using specific primers at the ends of each contig and ABI sequencing of the resulting PCR amplicons (Source Bioscience, Cambridge, UK). Sequences of the orfX region in S. sciuri isolate GVGS2 were submitted to the EMBL database under the accession number HG515014. Annotation was carried out using the automated RAST server28 and then manually with Artemis.29 Orthologous proteins were checked against the NCBI or EBI databases using BLAST. Comparative genomics was carried out using WebACT30 and viewed with the Artemis comparison tool (ACT).31 The presence of antibiotic resistance genes was identified using the ResFinder-1.3 Server (http://cge.cbs.dtu.dk/services/ResFinder/)32 and by BLAST. Nucleotide sequences of mecA homologues were aligned using ClustalW in Seaview33 and a maximum likelihood tree was generated using RAxML.34
PCR for SCCmec excision
Primers were designed using Primer 3 (http://primer3.sourceforge.net). Genomic DNA was extracted using the MasterPure Gram Positive DNA Purification Kit (Cambio, UK) from stationary-phase cultures grown in TSB. PCR was carried out using MyTaq DNA Polymerase (Bioline, UK). Primer sequences are listed in Table 2. PCR amplicons were ABI sequenced (Source Bioscience, Cambridge, UK).
Table 2.
Oligonucleotide primers used in this study
| Primer name | Sequence 5′–3′ | Target/function | Source |
|---|---|---|---|
| P1 | TATCATCGGCGGATCAAACG | detection of SCCmec excision | this work |
| P2 | TGCGGAGGCTAACTATGTCA | detection of SCCmec excision | this work |
| P3 | TTGCCAATTAAAAGGTTGGTTAG | detection of SCCmec excision | this work |
| P4 | TCTCAAGTAACATCTCAGCAATGA | detection of SCCmec excision | this work |
| P5 | TGTGGTGCCAATGTCAAAGT | detection of SCCmec excision | this work |
| P6 | TCGCTTTACAAGTGTCATGTTT | detection of SCCmec excision | this work |
| MecA1 | GTAGAAATGACTGAACGTCCGATAA | mecA | 52 |
| MecA2 | CCAATTCCACATTGTTTCGGTCTAA | mecA | 52 |
| mecC-Uni-F | GGATCTGGTACAGCATTACAACC | mecC/mecC1 | this work |
| mecC-Uni-R | TGCTTTAAATCRATMTTGCCG | mecC/mecC1 | this work |
| mecA1-spec-F | TTGAAGAAGCAACAACGCAC | mecA1 | this work |
| mecA1-spec-R | GAACCGTAGTCATCTTTCATGTTG | mecA1 | this work |
| Uni-16s-Ctrl-F | ACACGGTCCAGACTCCTACG | 16S rDNA | this work |
| Uni-16s-Ctrl-R | ATAATTCCGGATAACGCTTGC | 16S rDNA | this work |
Oligonucleotide primer design and strain screening
The sequences of mecC from S. aureus LGA251 and S. sciuri GVGS2 and mecC1 from S. xylosus S04009 were aligned with Seaview33 and conserved primers were designed using Primaclade.35 The presence of mecC was confirmed by PCR on boilates or genomic DNA using primers: mecC-Uni-F and mecC-Uni-R. Primer sequences are listed in Table 2. Boilates were prepared by inoculating two or three single colonies in 50 μL of sterile H2O and boiling for 5 min, followed by centrifugation at 16 000 g for 2 min.
Transcriptional analysis of mec gene expression by RT–PCR
Isolates GVGS2 and GVGS3 were grown in 5 mL of TSB supplemented with 0.1 mg/L oxacillin overnight at 37°C with 200 rpm shaking. After ∼16 h, the cultures were diluted 1/50 into 5 mL of fresh TSB supplemented with 0.1 mg/L oxacillin and grown for 3 h under the same conditions to an optical density of ∼0.8 at 595 nm. An S. sciuri mecA/mecA1-positive isolate and an ST130 S. aureus mecC-positive isolate were also grown under the same conditions as controls. Total RNA was then extracted from 1 mL of culture using the SV Total RNA Isolation System (Promega, UK) following the manufacturer's standard protocol for Gram-positive bacteria. After an additional DNAse step using RQ1 RNase-Free DNase (Promega, UK), cDNA was synthesized using ProtoScript® II Reverse Transcriptase (NEB, UK) and a Random Hexamer primer (Fisher Scientific, UK) following the manufacturer's standard protocol. Controls without reverse transcriptase were generated for all samples and showed no amplification in the subsequent PCRs. cDNA was used undiluted in a standard PCR for the detection of mecC (mecC-Uni-F/R), mecA (MecA1/A2) and mecA1 (mecA1-spec-F/R) (Table 2). PCR was carried out using MyTaq DNA Polymerase (Bioline, UK). A PCR for 16S rRNA (Uni-16s-Ctrl-F/R) was also carried out as a positive control for cDNA synthesis (Table 2).
Results
Multidrug-resistant S. sciuri subsp. carnaticus from wound infections in cattle
A farm in the south-west of England had multidrug-resistant bacterial infections in caesarean incision wounds in several Belgian Blue cattle. Multidrug-resistant Staphylococcus species (Table 1) were isolated from wound swabs taken from two cows (GVGS2 and GVGS3); both isolates were subjected to whole-genome sequencing. Analysis of 16S rRNA genes revealed these isolates to be S. sciuri. Further sub-speciation by BLAST comparison of the hsp60, sodA, dnaJ and tuf genes against the NCBI database identified the isolates as S. sciuri subsp. carnaticus.36,37 BLAST comparison of the four largest contigs (total size of contigs: 703 911 bp, ∼26% of GVGS2 genome) of the complete GVGS2 de novo genome assembly against GVGS3 identified only one single-nucleotide polymorphism (SNP), suggesting that the two isolates were very closely related (the same strain). The two isolates were resistant to a range of antimicrobial drugs (Table 1). Analysis of the genome sequence identified a number of resistance genes, including str, erm(C) (GVGS2 only), fexA and tet(K). These findings match the phenotype for these isolates, except for isolate GVGS2, which was susceptible to erythromycin on disc testing despite being positive for erm(C) (Table 1). Further analysis of the GVGS2 erm(C) gene revealed it to be part of a putative ∼2.5 kb plasmid (data not shown). The erm(C) gene was intact, but contained an Ile123Val substitution compared with the most closely related S. aureus erm(C) sequences in the NCBI database (accession number YP_001901404).
The orfX region of isolate GVGS2 contains both mecA and mecC
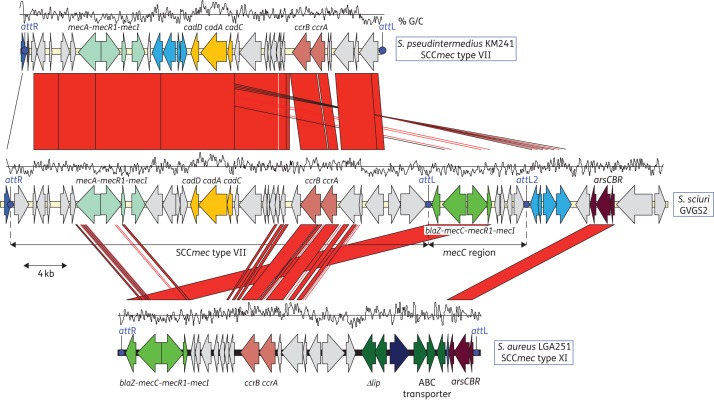
BLAST analysis identified that both isolates (GVGS2 and GVGS3) harboured three different homologues of the mecA gene: mecA, mecA1 and mecC. We further analysed the genome of GVGS2 in detail and identified that two of the mecA homologues (mecA and mecC) were found at the orfX locus (the SCCmec insertion site) (Figure 1), while mecA1 was part of the previously reported chromosomal locus that shared the greatest similarity to S. sciuri subsp. carnaticus strain ATCC 700058 (accession number AB547236) (data not shown).12 Comparative genomics of the orfX locus identified that the region was made up of two distinct parts; immediately downstream of the orfX locus was an SCCmec element that is most closely related to the SCCmec type VII in Staphylococcus pseudintermedius strain KM241 (Figure 1).38 The SCCmec in GVGS2 differed from the SCCmec type VII in S. pseudintermedius by the presence of a number of extra genes and a small deletion. Firstly, an extra hypothetical protein and a putative short-chain dehydrogenase/reductase were present at the 5′ end proximal to orfX and downstream of the ccrB5 gene, respectively, both of which are absent in S. pseudintermedius. Next, the two small hypothetical proteins present upstream of the ccrA gene in the S. pseudintermedius SCCmec were absent in GVGS2. At the 3′ end of the SCCmec, an extra AAA superfamily ATPase and a putative serine protease were also present in S. sciuri. The SCCmec element was bounded by two intact repeats (SCCmec attR and attL) (Figures 1 and 2). The region containing the mecC gene was immediately downstream of the SCCmec element and was bounded by a second SCCmec attL site at the 3′ end (attL2) (Figures 1 and 2). The mecC gene, as in S. aureus and S. xylosus, was part of a homologous class E mec gene complex (mecI-mecR1-mecC-blaZ).18,24 The mecC gene in GVGS2 shared 96.3% nucleotide identity with mecC from LGA251 and 91% nucleotide identity with mecC1 from S. xylosus. The other genes, mecI, mecR1 and blaZ, shared 95.6%, 97.1% and 97.7% nucleotide identity, respectively, with their respective homologues in LGA251. Four other genes were present between the mec gene complex and attL2. Immediately downstream of mecI was an AsnC family transcriptional regulator and putative glyoxalase, which were most closely related to an AsnC family transcriptional regulator in Clostridium arbusti SL206 (accession number ZP_10773559) and a glyoxalase/bleomycin resistance protein in Paenibacillus sp. JDR-2 (accession number YP_003008991), respectively. Next, there was a PhnB-like protein and a DeoR family putative transcriptional regulator, which are found in a number of S. aureus SCCmec elements in the DDBJ/EMBL/GenBank databases. Immediately downstream of the mecC region attL2 were four genes also present in the full SCCmec VII element upstream: a putative transmembrane protein, a putative transcriptional regulator, a rhodanese domain-containing protein and a metallo-β-lactamase superfamily protein. These genes are also part of the S. fleurettii chromosomal mecA locus that has been suggested to be the template for the mec complex in mecA SCCmec elements.12 Finally, downstream of this was an arsenic resistance gene cluster, arsCBR, which is part of the SCCmec type XI and which was also found to be present in the chromosome of S. xylosus S04009.24
Figure 1.
Comparison of the novel hybrid SCCmec-mecC in S. sciuri isolate GVGS2 (EMBL accession number HG515014), SCCmec type VII in S. pseudintermedius strain KM241 (EMBL accession number AM904731) and SCCmec type XI in S. aureus LGA251 (EMBL accession number FR821779). Areas of red show regions conserved between the two sequences and homologous coding DNA sequences are marked in the same colour. Blue dots indicate the SCCmec att sites. The percentage G/C content of the region is shown above each genome schematic.
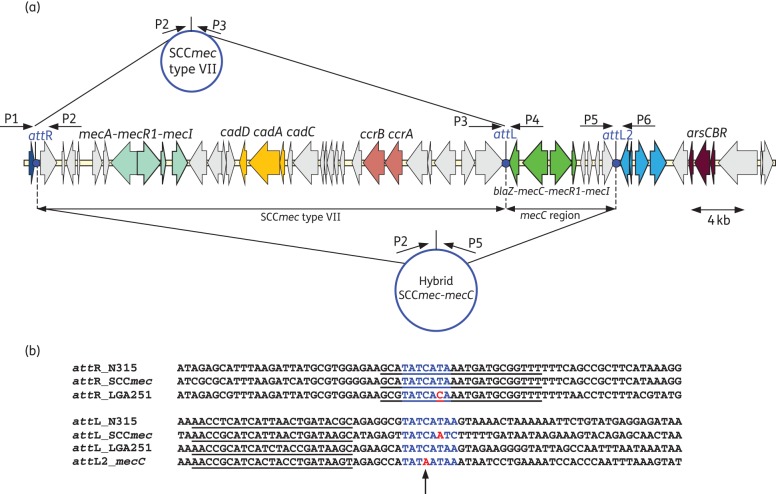
Figure 2.
SCCmec-mecC element excision and repeats. (a) Schematic representation of potential excised circular SCCmec and hybrid SCCmec-mecC, and location of PCR primers used to detect excision. (b) DNA sequences of attR and attL sites in S. aureus N315 (N315), SCCmec type VII in S. pseudintermedius strain KM241 (SCCmec), SCCmec type XI in S. aureus LGA251 (LGA251) and downstream of the mecC region in S. sciuri GVGS2 (mecC). The bases that make up the inverted repeat are underlined. The bases in blue represent the core 8 bp regions identified in the attB site with mutations highlighted in red.41 The central cytosine is thought to be essential for attB × attSCC recombination and is highlighted with an arrow.41
Both the SCCmec type VII and a hybrid SCCmec-mecC can excise from the chromosome
Previously, excision of a tandem arginine catabolic mobile element (‘ACME’)-SCCmec and a SCCmec type IV cassette at a secondary attR site (attR2) was reported in S. aureus.39,40 Further analysis of the flanking repeats showed that the attL2 repeat downstream of the mecC region contained an SNP (C to A) at the central cytosine previously shown to be essential for recombination between attB and attS (attSCC), suggesting that this repeat might not be functional (Figure 2b).41 The attR of the SCCmec also contained an SNP in the central 8 bp region in comparison with the attR of S. aureus N315 (T to A); however, substitutions in this position have been demonstrated not to adversely affect recombination.41 Therefore, as the SCCmec and mecC region in GVGS2 are bounded by a single attR and two different attL sites (attL and attL2) (Figures 1 and 2a) we designed PCR primers in order to detect excision and circularization of either the SCCmec type VII element alone (attR × attL) or a putative larger hybrid SCCmec-mecC element (attR × attL2) (Figure 2a). PCRs were designed to amplify across the orfX attB region if excision of either the SCCmec type VII alone (P1 + P4) or a hybrid SCCmec-mecC (P1 + P6) element occurred. A second set of PCRs were carried out to detect the putative extrachromosomal circular forms of either the SCCmec type VII (P2 + P3) or the SCCmec-mecC (P2 + P5) hybrid (Figure 2a). PCR conducted on ∼250 ng of genomic DNA from stationary-phase cultures produced weak positive PCR amplicons for P1 + P4, P1 + P6, P2 + P3 and P2 + P5 primer combinations. Sequencing of the PCR amplicons confirmed formation of attB between both attR × attL (P1 + P4) and between attR × attL2 (P1 + P6). Sequencing also confirmed the formation of the attSCC (present in the circular form) between attR × attL of the SCCmec type VI (P2 + P3) and the attR × attL2 of the hybrid SCCmec-mecC (P2 + P5).
Transcriptional analysis of mecC and mecA
In order to assess if both mecC and mecA were expressed in the same isolate, S. sciuri GVGS2 and GVGS3 were subjected to transcriptional analysis in the presence of low levels of oxacillin (0.1 mg/L). RT–PCR for mecC and mecA confirmed that both genes were expressed in GVGS2 and GVGS3 under the conditions tested, while no mecA1 transcript was detected.
Screening of S. sciuri isolates for mecC
Using a multiple sequence alignment of mecC from S. aureus LGA251, S. xylosus S04009 and S. sciuri GVGS2, we designed universal mecC primers and tested a selection of S. sciuri isolates to determine the prevalence of mecC genes. We tested 11 isolates of S. sciuri subsp. carnaticus isolated between 1990 and 1992 from different hosts (cattle, rodents and cetaceans) in the USA42 and 12 isolates from human clinical infections in England sent to Public Health England for further testing between 2006 and 2011. None of the isolates were positive by PCR for mecC.
Discussion
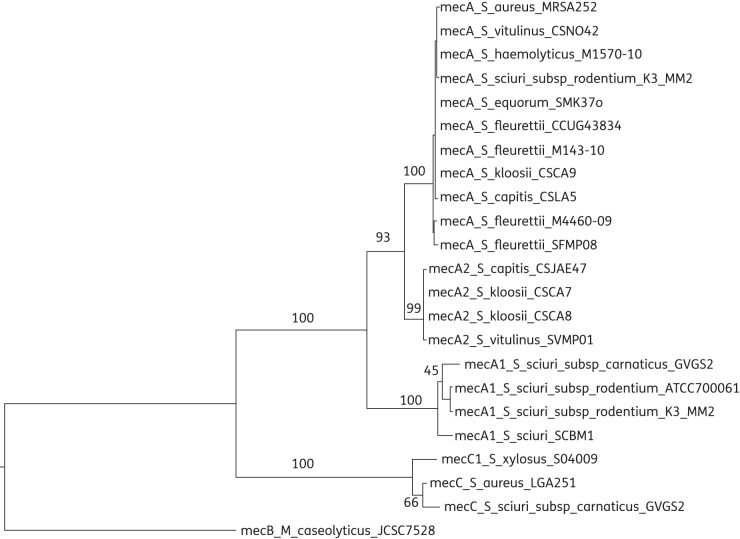
In this work, we have identified a further staphylococcal species that harbours the mecC gene. The mecC from GVGS2 is more closely related to mecC from S. aureus than mecC1 from S. xylosus. Phylogenetic analysis of mec gene homologues shows that the S. xylosus mecC1 probably represents a more ancestral form of mecC, as previously suggested (Figure 3).24 Like both S. aureus LGA251 and S. xylosus S04009, the S. sciuri isolates harbouring the mecC gene were again obtained from a bovine host, suggesting that selective pressure for the maintenance of mecC might be present in this or a closely linked ecological niche. mecC was also recently identified in a Staphylococcus stepanovicii isolate from a wild Eurasian lynx (Lynx lynx), suggesting that mecC-positive staphylococci are also present in diverse wildlife populations, as reported for S. aureus mecC isolates.16,43,44 We found that both mecC and mecA were expressed under laboratory growth conditions with low levels of oxacillin, suggesting that they may both contribute to the resistance phenotype of these isolates. The presence of both mecA and mecC in a single isolate is interesting, and suggests that the PBP2a proteins encoded by mecA and mecC might have distinct biological roles. This is further corroborated by the recent finding of a difference in temperature and substrate specificity of PBP2a encoded by mecC in comparison with mecA.45 It is of interest to find out how the two mec systems are regulated—whether regulation is hierarchal, with one system regulating the other, as seen with BlaR1/MecR1 regulation of mecA, and whether the recently described mecR2 also regulates mecC.46,47 Understanding the regulation of mecA and mecC under different conditions might provide further insights into the biology of the two mec genes and identify suitable measures for reducing the selective pressures that maintain them.
Figure 3.
Phylogenetic relationships of mec homologues. Maximum likelihood tree of nucleotide sequence of mec homologues. The tree is rooted in M. caseolyticus mecB as an outgroup. Bootstrap values for branches are shown.
We also identified that both the SCCmec type VII element and the SCCmec-mecC hybrid can excise from the chromosome and form a circular intermediate, despite the presence of SNPs in the attL2 and attR repeats. The fact that the C to A mutation in the attL2 does not prevent the excision reaction, as previously reported for the attB × attSCC integration reaction, suggests that this base is either not required for attL × attR recombination or that CcrA1 and CcrB5 have different sequence specificity compared with CcrA2 and CcrB2 from S. aureus N315 (71% and 86% amino acid identity, respectively). It is not possible to deduce if mecA and mecC were transferred together or independently into GVGS2 and GVGS3. There are no further regions of homology to either the SCCmec type XI or to the mecC region in S. xylosus S04009, which suggests that the mecC region was either transferred into the strain on a distinct element or has undergone significant decay. Recently, it was demonstrated that CcrA and CcrB recombinases can mediate recombination reactions between any combination of SCCmec repeats (attR/attL/attB/attSCC), raising the possibility that SCCmec type VII integrated into the attR of the mecC region or vice versa.7 The four genes immediately downstream from the attL2 of the mecC region are also present in the SCCmec VII element upstream and in the S. fleurettii chromosomal mecA locus, which has been suggested to be the template for the mec complex in mecA SCCmec elements (Figure 1).12 It is possible that these genes were also part of another SCCmec element that brought the mecC region into the chromosome. However, given that these genes are located outside of the mecC region attL2, it is equally likely that this just represents another, now decayed, SCC element present at the orfX locus.
The discrepancy of the presence of erm(C) and the lack of erythromycin resistance in GVGS2 is puzzling. The amino acid substitution in Erm(C) is unlikely to have caused a loss of function, as the Ile123Val mutation is present in a variable region of Erm-family proteins.48,49 A previous study has reported S. aureus erm(C)-positive isolates susceptible to erythromycin that could be selected to produce a resistance phenotype.50 Further investigation is required to understand the erythromycin-susceptible phenotype in GVGS2. In conclusion, this study further highlights that the mecC gene, like mecA, is disseminated widely amongst members of the Staphylococcus genus.
Nucleotide accession numbers
The nucleotide sequences determined for GVGS2 were deposited in the EMBL database under accession number HG515014.
Funding
This work was supported by a Medical Research Council Partnership Grant (G1001787/1) held between the Department of Veterinary Medicine, University of Cambridge (M. A. H.), the School of Clinical Medicine, University of Cambridge (S. J. P.), the Moredun Research Institute (R. N. Z.) and the Wellcome Trust Sanger Institute (J. P. and S. J. P.). S. J. P. receives support from the NIHR Cambridge Biomedical Research Centre. X. B. was supported by the China Scholarship Council and the Cambridge Overseas Trust. J. R. was supported by fellowship SFRH/BD/72675/2010 from Fundação para a Ciência e a Tecnologia.
Transparency declarations
Competing interests: none to declare.
The funder had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We would like to thank the George Veterinary Group for providing isolates, the core sequence team at the Sanger Institute for sequencing of the isolates described in this study, and Wesley Kloos for providing some of the S. sciuri subsp. carnaticus isolates that were used in this study.
References
- 1.Lim D, Strynadka NC. Structural basis for the β-lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol. 2002;9:870–6. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 2.Ubukata K, Nonoguchi R, Matsuhashi M, et al. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J Bacteriol. 1989;171:2882–5. doi: 10.1128/jb.171.5.2882-2885.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuda C, Suvorov M, Vakulenko SB, et al. The basis for resistance to β-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J Biol Chem. 2004;279:40802–6. doi: 10.1074/jbc.M403589200. [DOI] [PubMed] [Google Scholar]
- 4.Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984;158:513–6. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–55. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Archer GL. Roles of CcrA and CcrB in excision and integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol. 2010;192:3204–12. doi: 10.1128/JB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misiura A, Pigli YZ, Boyle-Vavra S, et al. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol Microbiol. 2013;88:1218–29. doi: 10.1111/mmi.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berglund C, Soderquist B. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden – possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin Microbiol Infect. 2008;14:1048–56. doi: 10.1111/j.1469-0691.2008.02090.x. [DOI] [PubMed] [Google Scholar]
- 9.Bloemendaal AL, Brouwer EC, Fluit AC. Methicillin resistance transfer from Staphylococcus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One. 2010;5:e11841. doi: 10.1371/journal.pone.0011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wielders CL, Vriens MR, Brisse S, et al. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet. 2001;357:1674–5. doi: 10.1016/s0140-6736(00)04832-7. [DOI] [PubMed] [Google Scholar]
- 11.Wu S, Piscitelli C, de Lencastre H, et al. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb Drug Resist. 1996;2:435–41. doi: 10.1089/mdr.1996.2.435. [DOI] [PubMed] [Google Scholar]
- 12.Tsubakishita S, Kuwahara-Arai K, Sasaki T, et al. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010;54:4352–9. doi: 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antignac A, Tomasz A. Reconstruction of the phenotypes of methicillin-resistant Staphylococcus aureus by replacement of the staphylococcal cassette chromosome mec with a plasmid-borne copy of Staphylococcus sciuri pbpD gene. Antimicrob Agents Chemother. 2009;53:435–41. doi: 10.1128/AAC.01099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walther B, Wieler LH, Vincze S, et al. MRSA variant in companion animals. Emerg Infect Dis. 2012;18:2017–20. doi: 10.3201/eid1812.120238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurent F, Chardon H, Haenni M, et al. MRSA harboring mecA variant gene mecC, France. Emerg Infect Dis. 2012;18:1465–7. doi: 10.3201/eid1809.111920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson GK, Larsen AR, Robb A, et al. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother. 2012;67:2809–13. doi: 10.1093/jac/dks329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuny C, Layer F, Strommenger B, et al. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One. 2011;6:e24360. doi: 10.1371/journal.pone.0024360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Alvarez L, Holden MT, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shore AC, Deasy EC, Slickers P, et al. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–73. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison EM, Paterson GK, Holden MTG, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. 2013;5:509–15. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petersen A, Stegger M, Heltberg O, et al. Epidemiology of methicillin-resistant Staphylococcus aureus carrying the novel mecC gene in Denmark corroborates a zoonotic reservoir with transmission to humans. Clin Microbiol Infect. 2013;19:E16–22. doi: 10.1111/1469-0691.12036. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Hiramatsu K, Tomasz A, et al. Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56:4997–9. doi: 10.1128/AAC.01199-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsubakishita S, Kuwahara-Arai K, Baba T, et al. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010;54:1469–75. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison EM, Paterson GK, Holden MT, et al. A Staphylococcus xylosus isolate with a new mecC allotype. Antimicrob Agents Chemother. 2013;57:1524–8. doi: 10.1128/AAC.01882-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miragaia M, Couto I, de Lencastre H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE) Microb Drug Resist. 2005;11:83–93. doi: 10.1089/mdr.2005.11.83. [DOI] [PubMed] [Google Scholar]
- 26.Quail MA, Kozarewa I, Smith F, et al. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–10. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz RK, Bartels D, Best AA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carver T, Harris SR, Berriman M, et al. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–9. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbott JC, Aanensen DM, Bentley SD. WebACT: an online genome comparison suite. Methods Mol Biol. 2007;395:57–74. [PubMed] [Google Scholar]
- 31.Carver TJ, Rutherford KM, Berriman M, et al. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 32.Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–63. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 35.Gadberry MD, Malcomber ST, Doust AN, et al. Primaclade – a flexible tool to find conserved PCR primers across multiple species. Bioinformatics. 2005;21:1263–4. doi: 10.1093/bioinformatics/bti134. [DOI] [PubMed] [Google Scholar]
- 36.Kloos WE, Ballard DN, Webster JA, et al. Ribotype delineation and description of Staphylococcus sciuri subspecies and their potential as reservoirs of methicillin resistance and staphylolytic enzyme genes. Int J Syst Bacteriol. 1997;47:313–23. doi: 10.1099/00207713-47-2-313. [DOI] [PubMed] [Google Scholar]
- 37.Zadoks RN, Watts JL. Species identification of coagulase-negative staphylococci: genotyping is superior to phenotyping. Vet Microbiol. 2009;134:20–8. doi: 10.1016/j.vetmic.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Descloux S, Rossano A, Perreten V. Characterization of new staphylococcal cassette chromosome mec (SCCmec) and topoisomerase genes in fluoroquinolone- and methicillin-resistant Staphylococcus pseudintermedius. J Clin Microbiol. 2008;46:1818–23. doi: 10.1128/JCM.02255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diep BA, Stone GG, Basuino L, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–30. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- 40.Jansen WT, Beitsma MM, Koeman CJ, et al. Novel mobile variants of staphylococcal cassette chromosome mec in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2072–8. doi: 10.1128/AAC.01539-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Safo M, Archer GL. Characterization of DNA sequences required for the CcrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol. 2012;194:486–98. doi: 10.1128/JB.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couto I, de Lencastre H, Severina E, et al. Ubiquitous presence of a mecA homologue in natural isolates of Staphylococcus sciuri. Microb Drug Resist. 1996;2:377–91. doi: 10.1089/mdr.1996.2.377. [DOI] [PubMed] [Google Scholar]
- 43.Loncaric I, Kubber-Heiss A, Posautz A, et al. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J Antimicrob Chemother. 2013;68:2222–5. doi: 10.1093/jac/dkt186. [DOI] [PubMed] [Google Scholar]
- 44.Monecke S, Gavier-Widen D, Mattsson R, et al. Detection of mecC-positive Staphylococcus aureus (CC130-MRSA-XI) in diseased European hedgehogs (Erinaceus europaeus) in Sweden. PLoS One. 2013;8:e66166. doi: 10.1371/journal.pone.0066166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim C, Milheirico C, Gardete S, et al. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J Biol Chem. 2012;287:36854–63. doi: 10.1074/jbc.M112.395962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arêde P, Milheiriço C, de Lencastre H, et al. The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of β-lactam resistance in MRSA. PLoS Pathog. 2012;8:e1002816. doi: 10.1371/journal.ppat.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hackbarth CJ, Chambers HF. blaI and blaR1 regulate β-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:1144–9. doi: 10.1128/aac.37.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maravic G, Bujnicki JM, Feder M, et al. Alanine-scanning mutagenesis of the predicted rRNA-binding domain of ErmC' redefines the substrate-binding site and suggests a model for protein-RNA interactions. Nucleic Acids Res. 2003;31:4941–9. doi: 10.1093/nar/gkg666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maravic G, Feder M, Pongor S, et al. Mutational analysis defines the roles of conserved amino acid residues in the predicted catalytic pocket of the rRNA:m6A methyltransferase ErmC'. J Mol Biol. 2003;332:99–109. doi: 10.1016/s0022-2836(03)00863-5. [DOI] [PubMed] [Google Scholar]
- 50.Martineau F, Picard FJ, Lansac N, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44:231–8. doi: 10.1128/aac.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Couto I, Wu SW, Tomasz A, et al. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J Bacteriol. 2003;185:645–53. doi: 10.1128/JB.185.2.645-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perez-Roth E, Claverie-Martin F, Villar J, et al. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001;39:4037–41. doi: 10.1128/JCM.39.11.4037-4041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]