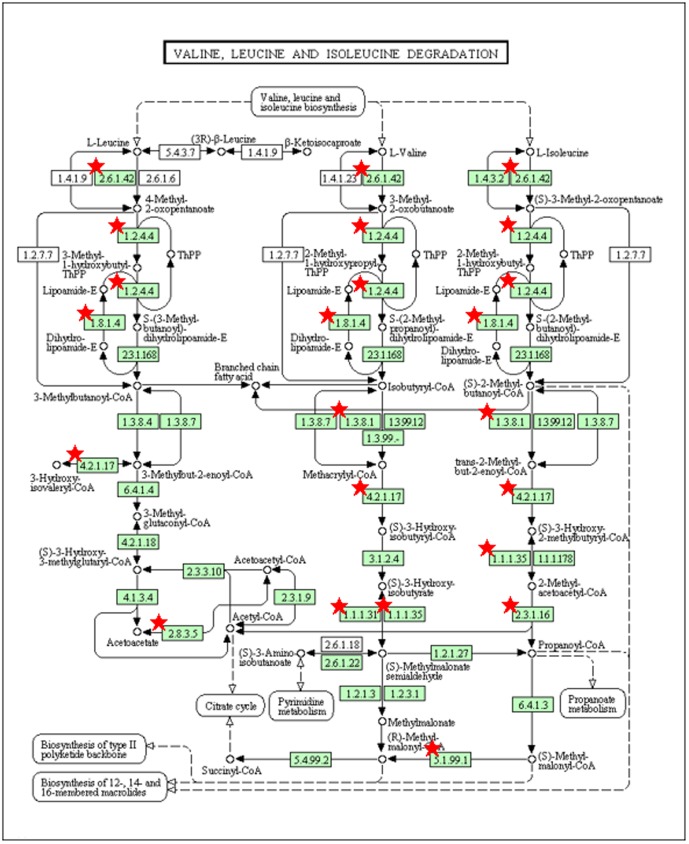
Figure 1. Pathway map of Val, Leu, and Ile degradation.
Individual gene name of the KEGG pathway of Val, Leu, and Ile degradation extracted by pathway analysis is shown as a metabolic map. Red asterisks indicate increased gene expression by microarray of Tg mice. Gene names corresponding to enzyme numbers with red asterisks are as follows: 2.8.3.5, 3-oxoacid CoA transferase 1; 2.3.1.16, acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase); 1.3.8.1, acyl-Coenzyme A dehydrogenase, short chain; 2.6.1.42, branched chain aminotransferase 2, mitochondrial; 1.2.4.4, branched chain ketoacid dehydrogenase E1, alpha polypeptide; 1.8.1.4, dihydrolipoamide dehydrogenase; 1.1.1.35, hydroxyacyl-Coenzyme A dehydrogenase; 4.2.1.17, hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit; 5.1.99.1, methylmalonyl CoA epimerase; 1.1.1.31, 3-hydroxyisobutyrate dehydrogenase.