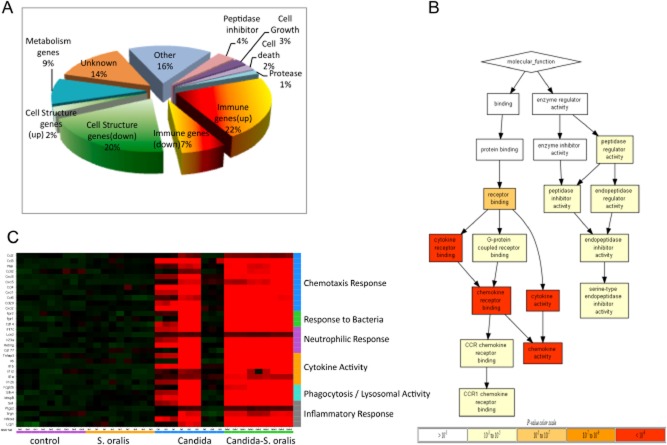
Fig 4.
C. albicans–S. oralis co-infection leads to a robust pro-inflammatory response. The oral transcriptional response to infection was analysed in tongue tissues from 3 animals per group on day 5 using whole mouse genome microarrays.A. Pie chart shows the representation of major functional categories of 195 genes differentially regulated in co-infected animals, compared with all other groups.B. molecular function tree of the differentially regulated genes in co-infected animals. The tree diagram displays the hierarchy of gene ontology terms and the levels of enrichment as indicated by FDR adjusted P-values. The genes used in the ontology analysis all exhibited greater than twofold change and adjusted P-value ≤ 0.05 in expression in the comparisons of the co-infected group against all other groups. Greater colour intensity indicates lower P-values.C. Heatmap showing relative expression levels of 31 selected genes across 4 groups of animals (3 biological replicates per group, each with technical triplicates). Normalization of each row (gene) for heatmap generation was performed by subtraction of the median value of the row followed with division by the median absolute deviation. Genes involved in similar biologic processes were grouped together in the categories shown on the right y-axis. Although there was variability in the Candida-infected animals, in the co-infected animals we found genes in the major categories of neutrophilic response/chemotaxis/inflammation to be consistently upregulated, indicative of a broad inflammatory response. Strong induction of multiple chemokines and other neutrophil-activating cytokines (e.g. IL-17C, TNF, IL-1α, IL-1β) were seen in mixed relative to single infection.