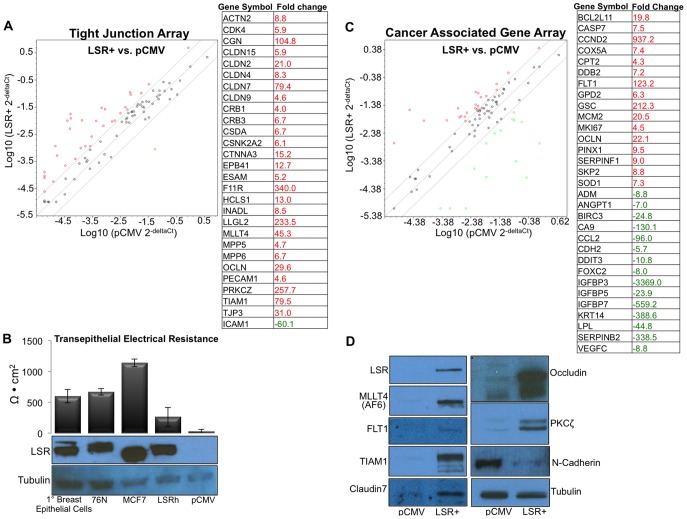
Figure 7. Re-introduction of LSR alters tight junction and breast cancer related gene expression.
Hs578t cells were stably transfected with either a control plasmid (pCMV), or a plasmid containing the full-length gene for LSR variant 1 (LSR+). Quantitative real-time PCR analysis was performed for genes associated and with tight junctions and adhesion proteins (A). Data show representative scatterplots and corresponding list of genes significantly altered by expression of LSR. (B) Transepithelial Electrical Resistance in breast epithelial cells and breast cancer cells +/− LSR expression. Cells were plated on transwell inserts and grown to confluence. Two days post formation of a high-density monolayer TER was measured directly in culture medium. Cell monolayers were directly lysed on the transwell insert following final TER measurement and subjected to western analysis using an LSR specific antibody. Tubulin was used as a loading control. Date represent a minimum of three independent experiments, each measured in triplicate. (C) Quantitative real-time PCR analysis was performed for genes associated and with deregulation in cancer. Data show representative scatterplots and corresponding list of genes significantly altered by expression of LSR. (D) Representative western blots highlighting protein level changes of a panel of genes found to be differentially expressed in the arrays.