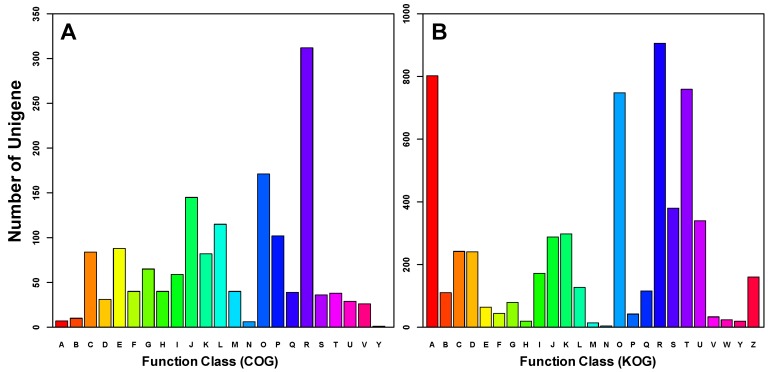
Figure 5.
Functional classification of differentially expressed transcripts between winged and wingless A. gossypii. Based on Clusters of Orthologous Groups (COG, Figure_5A), 676 genes (40.65% of the Differentially expressed transcripts ) were annotated and categorized into 23 functional classes. In comparison, 1144 genes (68.79% of the Differentially expressed transcripts ) were annotated and categorized into 25 specific classes within the euKaryotic Orthologous Groups (KOG, Figure_5B). A: RNA processing and modification, B: Chromatin structure and dynamics, C: Energy production and conversion, D: Cell cycle control, cell division, chromosome partitioning, E: Amino acid transport and metabolism, F: Nucleotide transport and metabolism, G: Carbohydrate transport and metabolism, H: Coenzyme transport and metabolism, I: Lipid transport and metabolism, J: Translation, ribosomal structure and biogenesis, K: Transcription, L: Replication, recombination and repair, M: Cell wall/membrane/envelope biogenesis, N: Cell motility, O: Posttranslational modification, protein turnover, chaperones, P: Inorganic ion transport and metabolism, Q: Secondary metabolites biosynthesis, transport and catabolism, R: General function prediction only, S: Function unknown, T: Signal transduction mechanisms, U: Intracellular trafficking, secretion, and vesicular transport, V: Defence mechanisms, W: Extracellular structures, Y: Nuclear structure, Z: Cytoskeleton.