Abstract
The National Heart, Lung and Blood Institute (NHLBI) is firmly committed to advancing translational research, especially in the field of genetics. An evaluation of the NHLBI’s extramural research grants funded in FY2008 and FY2011 was conducted to establish a baseline from which to assess progress in translational research, to assess current commitments and initial progress, and to identify putative gaps, barriers, and opportunities in the Institute’s human genetics research portfolios.
A search of the category of Genetics using the NIH Research, Condition, and Disease Categorization (RCDC) system was conducted to identify human genetics research project grants in the NHLBI’s genetics research portfolio. The NHLBI genetics portfolios were evaluated using a multidisciplinary research framework continuum that comprises five categories: discovery (T0); characterization (T1); clinical utility (T2); implementation, dissemination and diffusion (T3); and population health impact (T4). The abstracts for the grants were evaluated independently by two reviewers with an adjudicator for discrepancies in coding. The majority of the grants in 2008 and 2011 were classified as T0 and T1 research, with only four grants classified as T2 and beyond.
The majority of genetics grants funded in 2008 and 2011 were in the T0 and T1 categories, although the proportion of grants in T0 actually increased in that period. NHLBI-initiated programs to address this inability to move beyond T1 translation research have yet to have an impact on grant-funded translational genetic research. Future genetics studies should be designed with an eye towards translation to help overcome this barrier.
Keywords: genetics, translational medicine, NHLBI/NIH
Introduction
Translational medicine is a major focus of the National Institutes of Health (NIH) research agenda; NIH Director Francis Collins identified it as one of five promising areas ripe for major advances that could reap substantial downstream benefits.1 Since the publication of the NIH research agenda in January 2010, an advisory panel for the NIH proposed the creation of a new center focused primarily on translational medicine. As a result, President Barack Obama signed a spending bill on December 2011, launching the National Center for Advancing Translational Sciences (NCATS).2, 3 The increased focus on translational medicine, the nexus between basic science, and clinical and population based research to improve health, comes at an opportune time, especially in the field of genetics. The completion of the Human Genome Project (HGP), in conjunction with other advances in technology such as exome capture and sequencing, has greatly increased expectations for therapeutic development and the promise of personalized medicine.
Several institutes of the NIH are heavily engaged in efforts to explore the potential for high impact genetics findings to accelerate personalized medicine and population health benefits. The National, Heart, Lung and Blood Institute (NHLBI) has committed significant resources to genetics research in the last decade by supporting a large portfolio of population-based genetics programs in diverse US populations.4 The NHLBI aims to leverage the insights garnered from the discoveries in the genetics field to advance its mission of providing global leadership for research, training, and education programs that promote the prevention and treatment of heart, lung, blood, and sleep disorders.5 In keeping with Dr. Collins’s commitment to increased focus on translational research/medicine, we performed an evaluation of the NHLBI’s extramural research grants in human genetics. We focused on grants with initial funding or competing continuations (renewals) in Fiscal Year (FY) 2008 and FY 2011 to establish a baseline from which to assess initial commitments in translational research, to assess progress, and to identify scientific gaps, barriers, and opportunities in the Institute’s human genetics research portfolios.
Methods
Portfolio Analysis
The continuum of multidisciplinary research developed by Khoury et al. was utilized as a framework for evaluation of the NHLBI genetics portfolio. This translational research continuum comprises five categories or phases (Appendix 1). T0, the discovery phase, encompasses basic genetic research (including association studies). T1 includes characterization, generalization, and early evaluation of discoveries including clinical validity; functional studies are also classified as T1 research. T2 includes research conducted to evaluate candidate applications and clinical utility. T3 consists of established practice guidelines and relates to implementation, dissemination and diffusion research. T4 involves research conducted to determine population health impact, surveillance, or outcomes research.6
A compelling reason to utilize this approach was the clearly developed framework it provided. In addition, this framework was recently applied by the National Cancer Institute (NCI) to assess the NCI grant portfolio funded in fiscal years (FY) 2007 and 2010.7,8 The NHLBI portfolio analysis differed slightly from the NCI analysis in terms of the funding years assessed and the criteria for inclusion of grants in the analysis. The NHLBI analysis utilized the translational research continuum to evaluate investigator-initiated grants at two time points: (1) when funding for genetics projects was initially increasing in pace (FY2008), and (2) closer to the present year (FY2011). The NCI analysis included projects consisting of proteomics and metabolomics research activities; the NHLBI analysis excluded these unless there was a clearly defined genetics component.
An internal NIH database and multiple publicly accessible applications software packages were utilized to identify and analyze human genetics research projects composing the NHLBI’s genetics research portfolio. The internal and publicly accessible applications provide the same information, but have different interfaces. The database for the publicly accessible NIH data is called the NIH Research Portfolio Online Reporting Tools (RePORT- http://report.nih.gov/); the query form for the database is called RePORTER or the NIH Research Portfolio Online Reporting Tools Expenditures and Results (http://projectreporter.nih.gov/).9
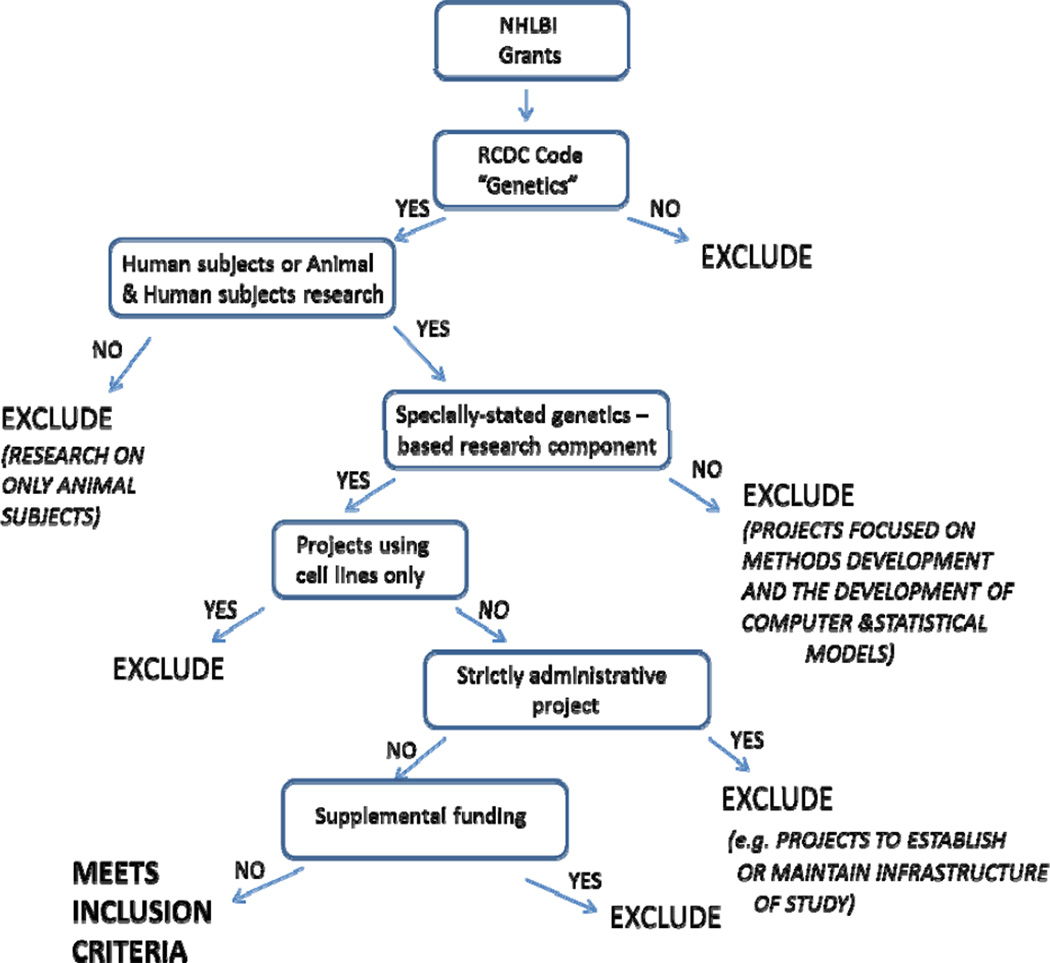
The internal web-based tool and RePORT integrate information from a database of information on extramural awards, database of financial obligations, and a database of indexed journals, citations, and abstracts. The Research, Condition, and Disease (RCDC) category of “Genetics” was queried for an initial screen of abstracts and titles of all grant applications receiving a competing award in FY2008 and FY2011.10 The RCDC System is an algorithm-based classifier that assigns codes, sorting NIH-funded projects into categories by research area, disease, or condition; an RCDC category is identical to the NIH Spending Category in RePORTER.11 The search results were subjected to a second review by the authors to ensure that the projects met the inclusion criteria for the analysis (see below). Figure 1 provides a graphical presentation of the inclusion and exclusion criteria.
Figure 1.
Inclusion & Exclusion Criteria Flowchart
Inclusion Criteria
To be included a research project had to meet all the following inclusion criteria:
Project assigned the RCDC Category of Genetics.
Projects that were classified as New and Competing Renewal in FY2008 or FY2011. (A New research project refers to a project/grant application that was not previously proposed or one that has not received prior funding; this is also known as Type 1. A Competing Renewals is a request for assistance to extend funding for one or more additional budget periods to continue a project for which funding would otherwise elapse; this is also known as Type 2.12)
Projects classified as research project grant or cooperative agreement (Appendix 2).
Projects conducted on human subjects. (If a project contained both animal and human research, the project was treated as human research.)
Projects that included a specifically-stated genetics-based research component.
Exclusion Criteria
Based on these criteria, the following types of projects were excluded from the analyses:
Projects that involved animal studies only.
Projects that focused on development of computer and statistical models.
Projects that focused on methods development.
Projects awarded for strictly administrative purposes such as to establish or maintain the infrastructure of a research study without providing funds to implement research protocols or conduct analyses. (e.g. core service labs, resequencing and genotyping centers, and DNA repositories).
Projects that involved research using cell lines only.
Projects representing supplemental funding (i.e. supplements to grants).
Each grant abstract was reviewed by non-overlapping pairs of reviewers working independently to determine if the project met the inclusion criteria for the portfolio analysis and, if so, to classify the research within the translational continuum. The projects were randomly divided into three collections and were evaluated and classified independently by reviewer pairs using the T0-T4 classification. For the grants with discrepant coding, the two reviewers jointly reviewed the abstracts to attempt to reach agreement. If no agreement was reached on the coding, a third reviewer, who was also one of the initial reviewers, served as an adjudicator in coding the grants. There was 75.7% agreement on the coding by the reviewers during the first review (340/449) and there was 85.5% (384/449) agreement on the coding during the subsequent review. It is important to note that abstracts of the grants were coded as the highest level of translational research proposed to be conducted by the grantee over the course of the 3–5 year grant period. For example, if T0, T1, and T2 work was proposed in a grant, the grant was coded as T2, even if that work would not be accomplished until the last year of the grant. Further, grants coded T2 or greater were re-evaluated independently by all reviewers (RF, MP, SS, PS, GP, or CJ) to ensure that the coding of grants in the more advanced stages of the translational continuum was consistent.
Results
Screening Results for Grants
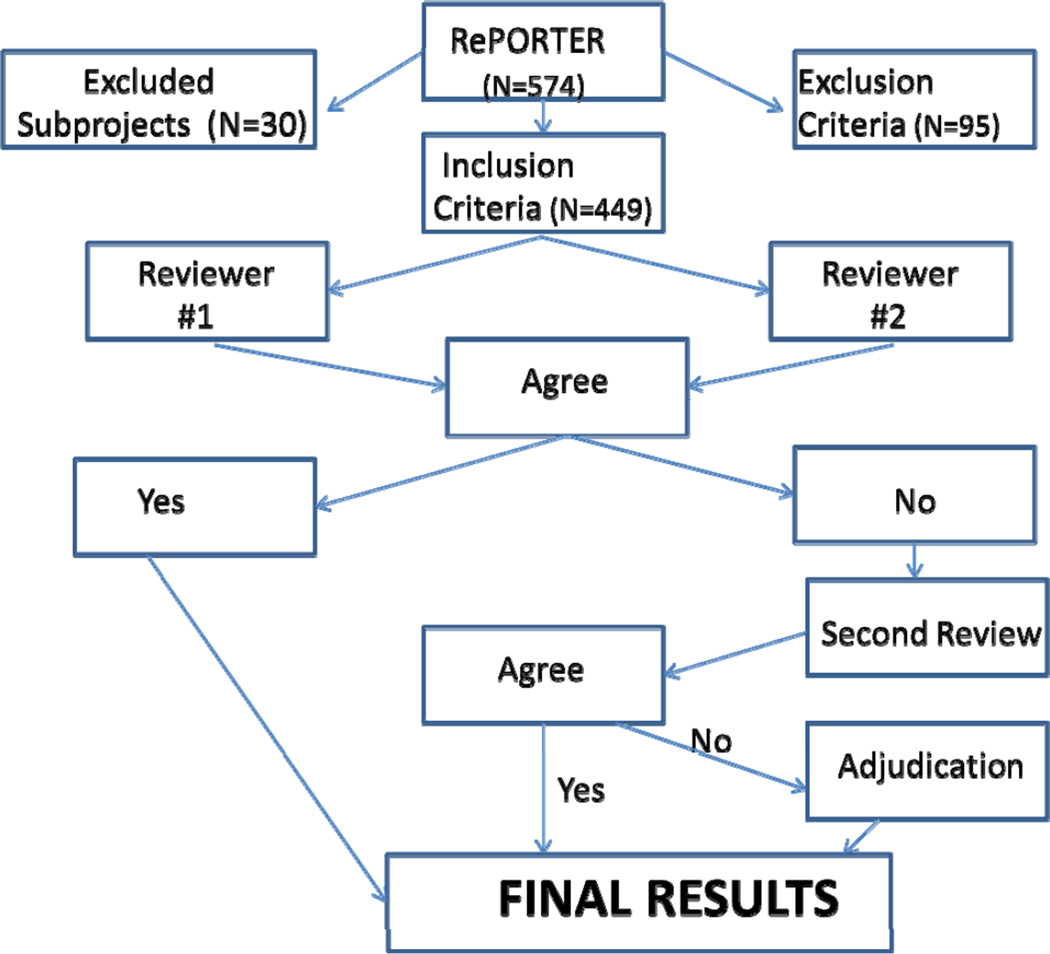
Figure 2 presents the results of the screening process for grants. The initial search results yielded a total of 574 grants. Several Program Project Grants (PPG/P01s) and Specialized Center (Cooperative Agreements/U54) included multiple subprojects; the codes for these grants were collapsed into one code to emphasize the highest level of translational research for that project. After staff review of abstracts, 16.5% of the grants were excluded from the analysis and 479 projects were determined to be eligible for inclusion in the portfolio analysis; of these 30 grants (6.3%) were excluded as they were subprojects to P01s and U54s that were only represented once in the portfolio analysis resulting in 449 grants being included in the portfolio analysis; this process avoids double-counting of grants. 250 grants were initially funded or had a competing continuation in FY2008 and 199 grants were newly funded or had a competing continuation in FY2011. The amount of funding for genetics grants was similar in FY2008 and FY2011, taking inflation into account. (NIH distributed $6,872,265,325 for genetics grants research in FY2008 and $7,223,000,000 in FY2011.9) This indicates that perhaps the grants are getting bigger as the level of funding holds steady.
Figure 2.
Screening Process for Review of Grants
Translation Classification Results for Grants
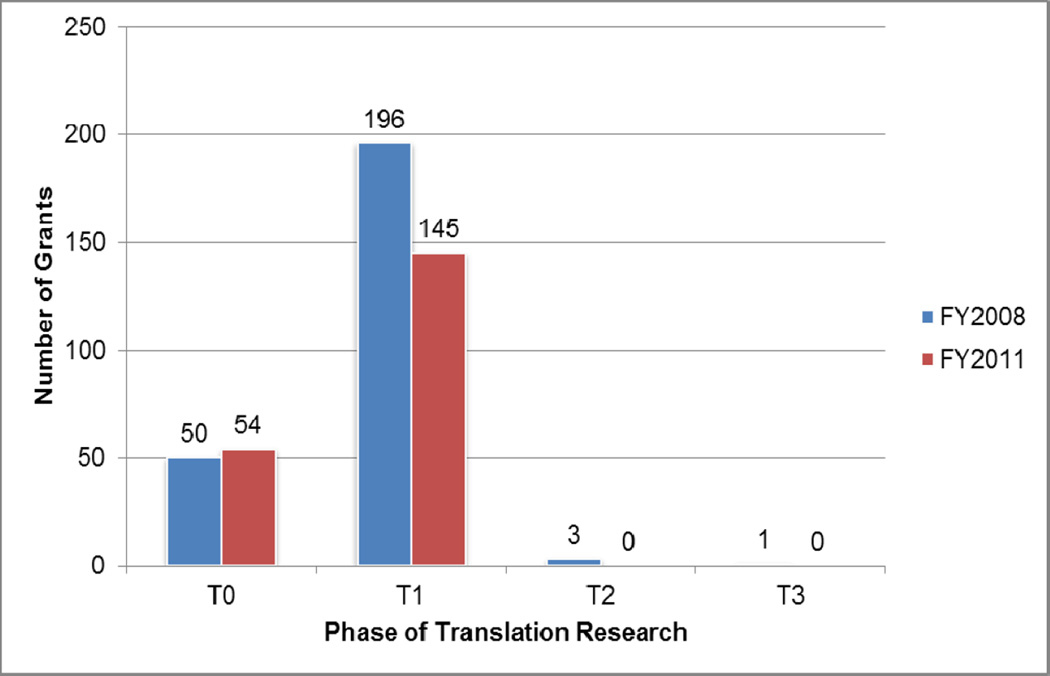
As seen in Figure 3, 20% of grants were included in the T0 phase, the majority (78.4%) in the T1 phase, and the remainder in the T2 and T3 phases (1.6% combined), for 2008. In 2011, 27.1% of the grants were coded T0, 72.9% were coded T1; no grants were coded T2 or later phase in this year. The analysis for indicates that the coded grants were distributed differently for FY2008 and FY2011. (Chi-square statistic = 6.92, p-value = .03)
Figure 3.
Translation Classification Results for Grants
Discussion
This portfolio analysis represents the first effort by NHLBI to evaluate its genetics research portfolio. The analysis was conducted on grants funded in 2008 and 2011, which characterizes the period before and after the implementation of the American Recovery & Reinvestment Act (ARRA).13 ARRA marked a significant increase in the amount of funding and number of research projects in genetics in 2009 and 2010. The results are similar to those of the NCI analysis in two ways. First, most of the genetics grants funded in 2008 and 2011 were in the T0 and T1 categories, and only a small number of grants are coded T2 or T3 and no grants are coded T4. Second, the proportion of grants in T0 actually increased from 2008 to 2011. The first finding of the analysis is not surprising as research suggests that it may take up to 17 years for genetics research to move down the translation pathway.14 Reasons for the first finding include: (1) Genetics technology for discovery has outpaced identification of functional variants and assessment of clinical validity and utility. ; (2) Functional studies require animal or cellular studies which can be difficult, as appropriate animal models may need to be developed and tissue samples from humans are not always accessible. Consequently there is a need to develop streamlined approaches to identifying functional variants, rare and common. ; (3) Clinical validity and utility assessments cannot be rushed immediately after functional As noted in a recent article, there is a necessary pause (for research) to get clinical validity and utility correct, lest we find ourselves spending money on technology with little benefit in the clinical setting. 15; (4) Larger sample numbers are required for statistically valid research that often require collaboration through consortia.16 The NHLBI has supported some of these consortia by use of NIH contractual arrangements rather than grants. ; and (5) Journal publication policies are evolving and the research findings now require replication and generalizability studies before publication.
The second finding that T0 grants actually increased in the later years is not surprising and again may reflect multiple causes: (1) Early genotyping technologies were not optimally designed to identify functional variants; (2) Genetic translation is not a linear process; and (3) Due to genetic heterogeneity, characterization and generalization to other populations may provide more evidence of false positives than replication of functional variants.
For all these reasons, the fact that T1 research has garnered and continues to garner the majority of genetic grant awards at this moment appears appropriate. Only four studies were classified as T2 or T3 research in 2008 and none in 2011 according to our analysis. The aims/objectives of the 2 studies coded as T2 research involved a pharmacogenetics study of warfarin response in blacks and a study of the pharmacogenetic effects of niacin on lipoproteins; the third involved the production and commercialization of an automated system to genotype and scan for variants associated with cystic fibrosis. The Warfarin study is part of the NHLBI supported Clarification of Optimal Anticoagulation through Genetics (COAG) trial. 17 An evaluation of physician communication about test results for newborn genetic screening represented the only grant coded as T3 research in our analysis. The NHLBI is responding to the dearth of grants classified as T2 to T4 by supporting various programs as indicated in the next section (The NHLBI Response).
The NHLBI Response
The NHLBI is funding programs/projects that (1) extend large-scale genotyping to non-European American populations, (2) evaluate gene interactions, (3) promote pharmacogenetics, (4) create new ways to obtain tissue- specific cells, and (5) conduct next generation sequencing. In addition, the NHLBI is involved with peripheral activities supportive of T2-T4 research.
Results of genotyping efforts in European American populations may not be generalizable to non-European populations, especially in regard to gene effect sizes and gene frequencies. The NHLBI is funding several large-scale genotyping efforts in non-European populations including the Candidate-gene Association Resource (CARe), which funded genotyping of African American participants from five of the nine cohorts including the Jackson Heart Study, an African American cohort; the study would be classified as T0 research.18 The NHLBI’s Omics in Latinos (Ola) program is another T0 research study that involves the genotyping and genetic analysis of non-European participants.19
The elucidation of gene by environment interactions is the focus of two programs supported by the NHLBI; these programs are classified as T1 research. The PROgram for Gene by ENvironment Interaction (PROGENI) aimed to identify novel gene by environment interactions by using short term, focused, interventions in families to identify genetic aspects of response to environmental changed and related biological mechanisms.20 The GEI: Genes, Environment, and Health Initiative program (partially funded by NHLBI) assessed gene by environment interactions. Innovative technologies were developed to assess environmental measures, dietary intake, and physical activity, and to determine a person’s biological response to these influences. The NHLBI supported the development of statistical methods to assess gene-environment interactions in complex diseases.21
Another high risk, high reward effort that can possibly result in personalized medicine is research in the field of pharmacogenetics, especially when an intervention/target has been identified. The National Institute of General Medical Sciences (NIGMS) sponsored Pharmacogenomics Research Network (PGRN) is a T1 research program designed to support research efforts to investigate genetic contributions to individual variability in drug therapy and the clinical utility of the findings by facilitating scientific collaborations.22 The NHLBI contributes support to the PGRN and the Pharmacogenetics Knowledgebase (PharmGKB), which integrates information obtained from the genotypes, phenotypes, and pharmacogenomics from PGRN, as well as other sources.23
The NHLBI’s Next Generation Genetic Association (Next Gen) Studies program represents a novel streamlined approach for identifying functional variants, through the use of induced pluripotent stem cells. Investigators funded through this T1 research program are required to utilize cellular reprogramming, molecular profiling, and genomics techniques to investigate how naturally occurring human genetic variation influences the activities of biological networks in cell-based models of disease.24 The resulting iPS cells from extensively genotyped and phenotyped participants will be made available to the scientific community as they are developed.
The NHLBI has also invested in whole exome and whole genome sequencing programs, as the results can help facilitate functional studies. The NHLBI Grand Opportunity Exome Sequencing Project (NHLBI GO-ESP), classified as T0 research, represents an effort to sequence the exomes of samples from NHLBI’s well-phenotyped populations.25 A recent publication by GO-ESP authors indicates that large sample sizes are required to associate rare variants with complex traits.26 However, family studies can achieve the same results by using smaller sample sizes. The Life After Linkage: The Future of Family Studies program (primarily funded by NHLBI) utilizes family studies for genetics research. The objective of this T1 research program is to integrate novel molecular data with existing genotype and phenotype data in families to identify and characterize genes influencing complex disorders through various methods including whole exome and whole genome sequencing. Findings in family studies might enable the discovery of additional and rarer variants and facilitate functional studies.27
NHLBI support of T2-T4 research
There are multiple examples of activities funded by NHLBI that facilitate T2-T4 research. The NHLBI is supporting efforts to identify biological pathways through the use of systems biology approaches and omics projects, such as proteomics and metabolomics. Systems approaches can be utilized to identify biological pathways and perhaps, new targets for interventions. The NHLBI Exploratory Program in Systems Biology, a T2 research program, applies systems biology approaches to innovative multidisciplinary research on the physiology and pathophysiology of heart, lung, blood, and sleep disorders.28
Omics projects can also be leveraged for identification of biological pathways that underlie pathophysiology of disease states. The NHLBI’s Anchoring Metabolomic Changes to Phenotype (P20) program aims to facilitate targeted metabolomic phenotyping studies on existing cohorts, population-based and family studies, intervention studies, and clinical studies. Aside from identifying metabolites and metabolomics profiles, the project seeks to uncover candidate pathways and genes responsible for the metabolites and metabolomics profiles of specific phenotypes; the program also hopes to identify, targets for intervention.29 The NHLBI Proteomics Centers employ proteomic technologies to understand physiologic pathways for defined clinical questions.30 Both of the programs mentioned above can be classified as T2 research.
Genomics is also embedded as part of some NHLBI training programs (T32 grants). Clinicians receive training in genomics that will be applicable to clinical diagnosis, treatment and prevention. By providing this sort of training NHLBI is preparing clinicians for implementation of genomic findings as they emerge.
Programs that were funded before or after the period of analysis were not included in the portfolio analysis. For example, the Cardiac Translational Research Implementation Program (C-TRIP) is not included, nor are the two components of the Bench to Bassinet Program, the Pediatric Cardiac Genetics Consortium (PCGC) and the Cardiovascular Development Consortium (CvDC); these programs are considered T2 research.31,32 The C-TRIP program uses a clinical trials approach to accelerate translation of new therapeutic interventions for treatment and prevention of heart failure or arrhythmias through execution of early-stage clinical efficacy trials. The mission of the Bench to Bassinet Program’s approach is to foster multidisciplinary collaborations to improve outcomes for people with congenital heart disease. Aside from the programs mentioned above, the NHLBI has convened several working groups to discuss pertinent issues in genetics research. In January 2009, a multidisciplinary working group was convened to update guidelines to the return of genetic research results, initially published in 2004.33 In August 2011, NHLBI hosted a working group to address issues regarding the integration and display of genetic test results within medical records. The group offered seven desiderata for the integration of genomic and other high volume biomolecular data into EHRs.34 Both working groups were discussing topics that are classified as T3 research.
Strengths of Analysis
The portfolio analysis uses an existing framework for the translation continuum of genetic research to clearly define the status of translational research funded by NHLBI in 2008 and 2011. The analysis is very similar to the analysis done by NCI and only one year later in time frame. Both institutes used the same coding framework but a different screening approach and eligibility criteria. In addition, there is an overlapping author with the NCI analysis to ensure consistency of coding methodology; the overlapping author was an initial reviewer and took on the role of the adjudicator. The use of existing publicly available data from RePORT for initial query is also beneficial because, as it facilitates replication by non-NIH investigators. The search results from RePORT utilized a readily accessible “screen” of projects and the reviewers/authors read all project abstracts, applying the attached review criteria to verify that the projects resulting from the queries belonged in the analysis. Each abstract had two reviewers; this two level review for eligibility for the analysis increased sensitivity of the process. The reviewers also met regularly to discuss coding strategies and to confer on several grants with discrepant coding. Finally, the team approach with adjudication of discrepancies promoted consistency in coding.
Limitations of Analysis
This analysis focused entirely on the NHLBI grant portfolio and ignored the major contract programs where much of the genetic research is funded by NHLBI. The analysis was based on the abstract, not the complete grant application. There were limits to the specificity of RePORT; the reviewers/authors read the project abstracts to tease out projects that did not belong in the analysis. Some studies with genetic components might have been excluded because the abstracts did not mention the research or used terms not in the search function. Cellular analyses and other omics that do not directly involve genetic-based analyses were excluded from the study. For example, research using epigenetics, proteomics, and metabolomics were excluded unless the research was linked to a specific gene or genetic pathway. The RCDC category definitions are updated as science advances; consequently the results of searches conducted on the RCDC Category or NIH Spending Category of “Genetics” have the potential to change the results over time.35 Changes in the categorization of projects are expected to be small; the distribution of T0-T3 coded grants is unlikely to be affected.
Conclusion
The portfolio analysis identifies putative gaps, barriers, and opportunities in the NHLBI’s human genetics grants research portfolio. The institute has demonstrated its strong interest in translational research by supporting programs and funding opportunities for investigators. To date, this analysis suggests the recent NHLBI programs have not moved translational genetics grants research beyond the T1 category. Our evaluation of the human genetics grants research portfolio should help the NHLBI and the investigator community assess the ongoing impact of current and future programs for translational genetics research.
Acknowledgments
We thank Dr. M.J. Khoury for his assistance in planning at the onset of this analysis. The authors would also like to acknowledge Dr. Melissa Antman for her guidance on the use of NIH data for the analysis.
Funding Sources: The authors conducted the analysis as part of their salaried duties as federal government employees.
Footnotes
Disclosures: None
Reference List
- 1.Collins FS. Research agenda. Opportunities for research and NIH. Science. 2010;327:36–37. doi: 10.1126/science.1185055. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health. National Center for Advancing Translational Sciences. 1-14-2013. Web. 1-11-2013. Website: http://www.ncats.nih.gov/
- 3.NIH Office of the Director (OD) NIH establishes National Center for Advancing Translational Sciences. NIH Office of the Director (OD) and NIH Office of Communications, editor. NIH News. 12-23-2011. Web. 1-20-2012. Website: http://www.nih.gov/news/health/dec2011/od-23.htm.
- 4.O'Donnell CJ, Nabel EG. Cardiovascular genomics, personalized medicine, and the National Heart, Lung, and Blood Institute: part I: the beginning of an era. Circ Cardiovasc Genet. 2008;1:51–57. doi: 10.1161/CIRCGENETICS.108.813337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Institutes of Health. NHLBI Mission Statement. 6-1-2009. Web. 1-20-2012. Website: http://www.nhlbi.nih.gov/about/org/mission.htm.
- 6.Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9:665–674. doi: 10.1097/GIM.0b013e31815699d0. [DOI] [PubMed] [Google Scholar]
- 7.Schully SD, Benedicto CB, Gillanders EM, Wang SS, Khoury MJ. Translational research in cancer genetics: the road less traveled. Public Health Genomics. 2011;14:1–8. doi: 10.1159/000272897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schully SD, Benedicto CB, Khoury MJ. How can we stimulate translational research in cancer genomics beyond bench to bedside? Genet Med. 2012;14:169–170. doi: 10.1038/gim.2011.12. [DOI] [PubMed] [Google Scholar]
- 9.Office of Extramural Research U.S. Department of Health & Human Services: Research Portfolio Online Reporting Tools (RePORT). 2-28-2013. Web. 3-26-2013. Website: http://report.nih.gov/
- 10.Office of Extramural Research. The Research, Condition, and Disease Categorization Process. 5-16-2012. Web. 4-23-2013. Website: http://report.nih.gov/rcdc/
- 11.Office of Extramural Research. Estimates of Funding for Various Research, Condition, and Disease Categories (RCDC). 4-10-2013. Web. 4-26-2013. Website: http://report.nih.gov/categorical_spending.aspx.
- 12.National Institutes of Health, Office of Extramural Research. Grants & Funding: Glossary & Acronym List. 3-20-2013. Web. 4-29-2013. Website: http://grants.nih.gov/grants/glossary.htm#G.
- 13.U.S. Department of Health & Human Services. NIH and the American Recovery and Reinvestment Act (ARRA). 3-29-2013. Web. 4-15-2013. Website: http://recovery.nih.gov/
- 14.Westfall JM, Mold J, Fagnan L. Practice-based research--"Blue Highways" on the NIH roadmap. JAMA. 2007;297:403–406. doi: 10.1001/jama.297.4.403. [DOI] [PubMed] [Google Scholar]
- 15.Holmes MV, Perel P, Shah T, Hingorani AD, Casas JP. CYP2C19 genotype, clopidogrel metabolism, platelet function, and cardiovascular events: a systematic review and meta-analysis. JAMA. 2011;306:2704–2714. doi: 10.1001/jama.2011.1880. [DOI] [PubMed] [Google Scholar]
- 16.Ioannidis JP. This I believe in genetics: discovery can be a nuisance, replication is science, implementation matters. Front Genet. 2013;4:33. doi: 10.3389/fgene.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.COAG Research Network. Clarification of Optimal Anticoagulation through Genetics (COAG). 1-1-2009. Web. 2-10-2013. Website: http://coagstudy.org/
- 18.Musunuru K, Lettre G, Young T, et al. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.NIH/NHLBI. FedBizOpps.gov: Federal Business Opportunities -Omics in Latinos (OLa): Genetics Analysis. 9-12-2011. Web. 3-29-2013. Website: https://www.fbo.gov/index?s=opportunity&mode=form&id=616a240163cf25f7559730879a9e6a65&tab=core&_cview=1.
- 20.NIH/NHLBI. PROGENI: NHLBI Programs in Genes Environment Interactions Network, NHLBI.NIH. 9-01-2011. Web. 4-26-2013. Website: http://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/progeni.htm.
- 21.NIH/NHLBI. GEI: Genes, Environment, and Health Initiative, NHLBI. NIH. 9-01-2013. Web. 4-26-2013. Website: http://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/gei.htm.
- 22.NIH/NHLBI. PGRN: Pharmacogenomics Research Network, NHLBI. NIH. 9-01-2013. Web. 4-26-2013. Website: http://www.nhlbi.nih.gov/resources/geneticsgenomics/programs/pgrn.htm.
- 23.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S. Department of Health & Human Services. Next Generation Genetic Association Studies (U01): RFA-HL-11-006. 3-15-2010. Web. 3-12-2013. Website: http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-11-006.html.
- 25.NIH/NHLBI. NHLBI Grand Opportunity Exome Sequencing Project (ESP). 4-1-2013. Web. 4-15-2013. Website: https://esp.gs.washington.edu/drupal/
- 26.Tennessen JA, Bigham AW, O'Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.U.S. Department of Health and Human Services. Life After Linkage: The Future of Family Studies (R01); RFA-HL-12-007. 5-3-2011. Web. 4-16-2013. Website: http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-12-007.html.
- 28.U.S. Department of Health & Human Services. NHLBI Exploratory Program in Systems Biology (R33): RFA-HL-07-005. 6-5-2006. Web. 2-15-2013. Website: http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-07-005.html.
- 29.U.S. Department of Health & Human Services. Anchoring Metabolomic Changes to Phenotype (P20): RFA-HL-12-009. 4-5-2011. Web. 4-5-2013. Website: http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-12-009.html.
- 30.U.S. Department of Health and Human Services: National Heart LaBI. NHLBI Proteomics Centers: Translating Proteomics Knowledge and Tools to Advance Biology and Medicine. 4-25-2013. Web. 4-26-2013. Website: http://www.nhlbi-proteomics.org/
- 31.NIH/NHLBI. Bench to Bassinet Program: Supporting Excellence in Pediatric Cardiovascular Translational Research. 4-1-2013. Web. 4-15-2013. Website: http://www.benchtobassinet.com/
- 32.U.S. Department of Health & Human Services. Cardiac Translational Research Implementation Program (C-TRIP) (P20): RFA-HL-006. 10-29-2008. Web. 4-26-2013. Website: http://grants.nih.gov/grants/guide/rfa-files/RFA-HL-10-001.html.
- 33.Fabsitz RR, McGuire A, Sharp RR, Puggal M, Beskow LM, Biesecker LG, et al. Ethical and practical guidelines for reporting genetic research results to study participants: updated guidelines from a National Heart, Lung, and Blood Institute working group. Circ Cardiovasc Genet. 2010;3:574–580. doi: 10.1161/CIRCGENETICS.110.958827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masys DR, Jarvik GP, Abernethy NF, et al. Technical desiderata for the integration of genomic data into Electronic Health Records. J Biomed Inform. 2012;45:419–422. doi: 10.1016/j.jbi.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Institutes of Health. Reasons Funding Levels Might Change. 5-16-2012. Web. 5-14-2013. Website: http://report.nih.gov/rcdc/reasons.aspx.