Abstract
Morphogens are substances that establish a graded distribution and elicit distinct cellular responses in a dose dependent manner. They function to provide individual cells within a field with positional information, which is interpreted to give rise to spatial patterns. Morphogens can consist of intracellular factors that set up a concentration gradient by diffusion in the cytoplasm. More commonly, morphogens comprise secreted proteins that form an extracellular gradient across a field of cells. Experimental studies and computational analyses have provided support for a number of diverse strategies by which extracellular morphogen gradients are formed. These include free diffusion in the extracellular space, restricted diffusion aided by interactions with heparan sulfate proteoglycans, transport on lipid-containing carriers or transport aided by soluble binding partners. More specialized modes of transport have also been postulated such as transcytosis, in which repeated rounds of secretion, endocytosis and intracellular trafficking move morphogens through cells rather than around them, or cytonemes, which consist of filopodial extensions from signal receiving cells that are hypothesized to reach out to morphogen sending cells. Once the gradient has formed, cells must distinguish small differences in morphogen concentration and store this information even after the gradient has dissipated. This is often achieved by translating ligand concentration into a proportional increase in numbers of activated cell surface receptors that are internalized and continue to signal from endosomal compartments. Ultimately, this leads to activation of one or a few transcription factors that transduce this information into qualitatively distinct gene responses inside the nucleus.
Introduction
Pattern formation is the process by which a specialized cell, the fertilized egg, divides to give rise to seemingly identical cells. Over time and space, these cells take on distinct fates and eventually become organized into functional organ systems. Within a single organ, such as the heart, multiple classes of differentiated cells must synchronize their duties to ensure that the organ as a whole is functional. In addition, intricate coordination of all organ systems is required to generate a functional, mature animal. The complexity of this process is enormous, even at the level of the simplest invertebrate models and seems overwhelming in the case of higher vertebrates. While much remains to be understood about how an egg gives rise to the adult, one of the unifying principles of development is the idea that embryogenesis proceeds via the iterative process of generating naïve fields of cells, and then providing cells within each field with unique positional information, which they then interpret to give rise to spatial patterns. Through this process, the embryo is sequentially subdivided, initially along the major body axes, and then into smaller, and more refined units such as organ primordia that are further partitioned and patterned.
A fundamental problem in embryonic patterning is how naïve fields of cells are provided with positional information. Over 50 years ago, an elegant solution to this problem was hypothesized in the form of a morphogen gradient1–3. Morphogens are substances that form concentration gradients across fields of cells or nuclei and elicit distinct cellular responses in a dose dependent manner. Morphogens can consist of cytoplasmic proteins, such as transcription factors that form a gradient by diffusion within a single cell or syncytium, or secreted signaling molecules that travel from cell to cell. In most cases, morphogens guide the generation of different cell types in a specific spatial order, usually by inducing unique transcriptional responses in a dose dependent manner. More recent studies have shown that morphogen gradients can also provide positional information to organize cell polarity rather than specify cell fate within a field4. In this case the direct readout is independent of transcription. Morphogens also function to coordinate organ growth and patterning5.
The existence of morphogen gradients, and the identity of many morphogens are now well established. Although each gradient is distinguished by unique features, common strategies used to generate morphogen gradients can be identified. This article draws on selected classic examples from the literature to illustrate the major mechanisms by which morphogen gradients are hypothesized to be formed and fine-tuned, and how cells read their position within the gradient. It concludes with a brief description of the next level of systems organization, how gradients are interpreted.
ESTABLISHMENT OF INTRACELLULAR MORPHOGEN GRADIENTS
The first identified morphogen, and perhaps the best studied, is the transcription factor Bicoid6. Bicoid forms a gradient across the anterior-posterior axis of the early Drosophila embryo that patterns the head and thorax7, 8. The Bicoid gradient forms in an embryonic syncytium consisting of a common cytoplasm supporting many nuclei9. This is a relatively unique developmental context, and one in which the problem of morphogen distribution is simplified since a gradient can be established by simple diffusion from the source. Despite this seeming simplicity, the exact mechanism by which the Bicoid gradient forms remains controversial.
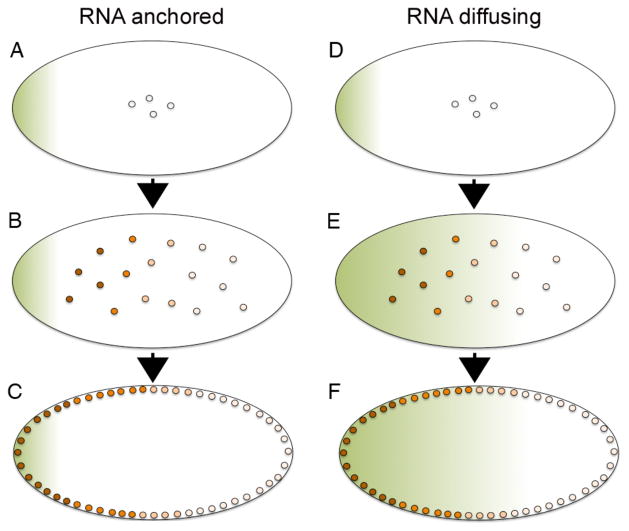
The Bicoid protein gradient originates with bicoid messenger RNA, which is deposited at the anterior pole of the fertilized egg10 (Figure 1, green shading). Following fertilization, the embryo undergoes 13 synchronous rounds of nuclear division that are not accompanied by cellular division. During the initial nuclear divisions, the egg is a homogeneous mixture of yolk granules and cytoplasm with nuclei distributed throughout the core (Fig. 1A,B,D,E), but during later cycles the nuclei move to the surface of the embryo (Fig. 1C,F). At approximately 3 hours of development, the embryo consists of ~6000 nuclei located in a cortical shell, surrounding the yolk in the center of the egg. A gradient of immunoreactive Bicoid protein can be detected within nuclei even prior to this time, with concentration decreasing as a function of distance from the anterior pole8 (Fig. 1B,C, orange shading). The primary mechanism by which Bicoid specifies cell fate in the early embryo is by activating or repressing transcription of its target genes, with different genes being transcribed at different locations in the embryo based on the concentration of bicoid protein present in the nuclei at that position7, 8, 11–14.
Figure 1. Illustration of the Bicoid gradient in the early syncitial Drosophila embryo.
(A–F) Following fertilization of the Drosophila egg, nuclei divide in the absence of cell division and remain positioned near the center of the embryo for the first 8 cell cycles (A–B, D–E) but then migrate to the periphery (C,F). bicoid RNA (green shading) is deposited at the anterior pole of the egg (A,D) and a nuclear Bicoid protein gradient (orange shading) forms either by translation of the localized RNA followed by protein diffusion away from the anterior pole (AC) or by diffusion of RNA away from the anterior pole followed by local translation of the graded RNA (D–F). Anterior is to the left in all panels.
The dynamics of Bicoid gradient formation have been analyzed using live imaging of green fluorescent protein (GFP) tagged forms of Bicoid15–17 and four models have been proposed to explain how the gradient forms18, with two of these predominating19. The most straightforward way in which this gradient might form is by localized translation of bicoid RNA at the anterior pole and simple diffusion of the protein away from the source, accompanied by a constant rate of degradation8, 20 (Fig. 1A–C). This model is widely accepted, although calculations based on experimental measurements suggest that the rate of diffusion of Bicoid protein is too slow to account for the observed dynamics of gradient formation17. Alternatively, it has been proposed that the Bicoid protein gradient is pre-configured by a bicoid RNA gradient21 (Fig. 1D–F). This model is attractive, since local translation of a graded RNA makes the issue of protein diffusion rates irrelevant for gradient formation. It is also consistent with the observation that bicoid RNA is released from the anterior pole following fertilization22, and that its distribution matches that of Bicoid protein at the time that a stable protein gradient can first be detected21. Careful analysis of the dynamics of bicoid mRNA gradient formation, however, suggests that the spatial distribution of bicoid RNA contributes to, but cannot fully account for the protein gradient, and that movement of both RNA and protein is essential23.
Bicoid provides an elegant, simple example of how morphogens can pattern embryos by bringing two predominant factors into play. First, the gradient forms in a unique developmental context where cell walls do not provide a barrier to free movement. Second, the morphogen itself directly induces changes in gene expression that specify distinct cell fates rather than relying on signals relayed from downstream receptors and cytoplasmic components to effect changes in gene expression. While this direct route to gradient formation is less common in vertebrates, there are a few examples of analogous mechanisms operating in early embryos. In Xenopus, for example, RNA encoding the transcription factor VegT is anchored to the vegetal cortex in the oocyte (Fig. 2A, green shading) but is released and becomes distributed throughout the vegetal half of the early cleavage stage embryo19, 24–26 (Fig. 2B). The RNA is then translated to generate a graded distribution of nuclear VegT protein that is restricted to prospective endodermal cells27 (Fig. 2B, C orange shading). During the late blastula stage, VegT activates transcription of a number of target genes including those encoding secreted morphogens of the transforming growth factor-β (TGF-β) family, such as nodal28, 29 (Fig. 2C, small white circles). Thus, in this example an initial transcription factor gradient is formed prior to cellularization, and the downstream targets of this transcription factor are extracellular proteins that further propagate the signal in the multicellular embryo30. This morphogen cascade is responsible for dose dependent induction of endodermal and mesodermal fates along the animal-vegetal axis31. Specifically, cells closest to the vegetal pole are exposed to a high concentration of nodal and adopt an endodermal fate, while those near the equator sense a lower dose of nodal and become mesoderm (Fig. 2C). Another example of a gradient that initially forms independent of protein movement between cells, is the fibroblast growth factor 8 (FGF8) gradient that forms in the presomitic mesoderm of early chick and mouse embryos32. In this case, an RNA gradient is established by restricted transcription of the fgf8 gene in the most immature cells at the posterior tip of the tail. As cells move away from the posterior pole and begin to mature, the fgf8 gene is no longer transcribed and existing transcripts slowly decay, setting up a posterior- to anterior- gradient of fgf8 mRNA, which is then translated to generate a corresponding protein gradient. This situation is similar to the bicoid mRNA gradient in Drosophila, but is also different in that the fgf8 gradient forms across a field of cells rather than within the confines of a single cell. Furthermore, the RNA is translated to make a secreted protein rather than a transcription factor, and this forms a secondary gradient that can move outside of cells.
Figure 2. The VegT-nodal morphogen gradient in the early Xenopus embryo.
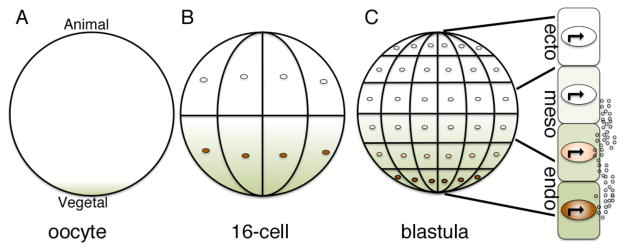
(A–C) vegT RNA (green) is anchored to the cortical cytoskeleton at the vegetal pole of the oocyte (A) but is released upon oocyte maturation and diffuses toward the animal pole during early cleavage stages (B). Local translation of vegT RNA generates a gradient of VegT protein (orange) that is restricted to nuclei of prospective endodermal cells located near the vegetal pole (B,C). VegT activates transcription of target genes such as nodal and other TGF-β family members, which encode secreted morphogens (C, small circles). Nodal is proposed to specify endodermal (endo) and mesodermal (meso) fate at high and moderate doses, respectively. Cells that are not exposed to nodal adopt an ectodermal (ecto) fate.
ESTABLISHMENT OF EXTRACELLULAR MORPHOGEN GRADIENTS
Intracellular morphogen gradients formed by diffusion and/or localized translation in the cytoplasm are the exception rather than the rule. A more common mechanism of gradient formation in development is the locally restricted transcription of a gene encoding a secreted protein, followed by movement of the signaling molecule across or through a field of cells. Many different secreted proteins, including members of the Wnt [Wingless (Wg) in Drosophila], Hedgehog (Hh), FGF and TGF-β families, have been shown to function as morphogens. In each case, a stable gradient of extracellular protein is organized across a field of cells. The amount of ligand present at each position determines the level of activation of the associated cell surface receptor and downstream intracellular signal transduction cascade, and this in turn triggers dose dependent activation or repression of distinct target genes.
At first glance, formation of an extracellular gradient seems to be a simple matter of free diffusion of the morphogen away from its source followed by degradation, either outside of receiving cells or following receptor activation and internalization. In reality, the problem is considerably more complex. High affinity interaction of ligands with their cognate receptors restricts free diffusion and strongly influences the shape of morphogen gradients. In addition to physically restricting movement, receptors play important roles in either stabilizing ligands on the cell surface33 or targeting them for endocytosis and lysosomal degradation34. Secreted ligands interact not only with signal transducing receptors, but also with numerous proteins present on the cell surface or in the extracellular matrix and these binding partners can hinder and/or enhance extracellular movement. It is difficult to isolate and assess the role of these binding proteins in ligand transport because they often have more direct functions in promoting or inhibiting intracellular signal transduction, which clouds interpretation of the effect of their removal on gradient formation. An additional complication arises in that some ligands are post-translationally modified by the covalent addition of lipids and thus remain tightly associated with the plasma membrane, a process that is seemingly incompatible with long-range extracellular movement. Because the various extracellular signaling molecules that work as morphogens have very different biophysical and biochemical properties there is no single mechanism that can fully explain gradient formation in all cases. It is likely, in fact, that different mechanisms are used by the same morphogen in different cellular contexts. Experimental evidence coupled with theoretical predictions supports three major mechanisms by which extracellular morphogen gradients can form and these are presented below.
Movement in the extracellular space
The best-studied mechanism by which secreted proteins establish a long-range extracellular gradient is by movement through the extracellular space. Direct evidence for this model was initially hard to obtain due to the difficulty of detecting small amounts of soluble proteins in the extracellular space against a background of intracellular protein that is either in transit to the cell surface during its biosynthesis, or that has been re-internalized following secretion. In recent years, technical improvements in antibody staining and the ability to visualize the real time movement of fluorescently tagged proteins in unfixed tissues have confirmed the role of extracellular movement in establishing morphogen gradients. In a few cases, morphogens appear to move by passive diffusion through the extracellular space. The TGF-β family members activin, nodal and TGF-β1, for example, appear to move relatively freely from their source to establish an extracellular gradient in Xenopus35, 36 tissue recombination experiments35, 36 (Fig. 3A). More often, however, morphogen movement is regulated by local interactions with other molecules in a process termed restricted or facilitated diffusion. Evidence supporting the role of heparan sulfate proteoglycans (HSPGs), lipid containing particles and soluble extracellular binding proteins in promoting diffusion of secreted proteins is discussed below. It should be kept in mind that different morphogens employ different binding partners, that a single morphogen may utilize more than one of these binding partners in different contexts or even in the same transport process, and that binding partners may function in processes other than ligand transport.
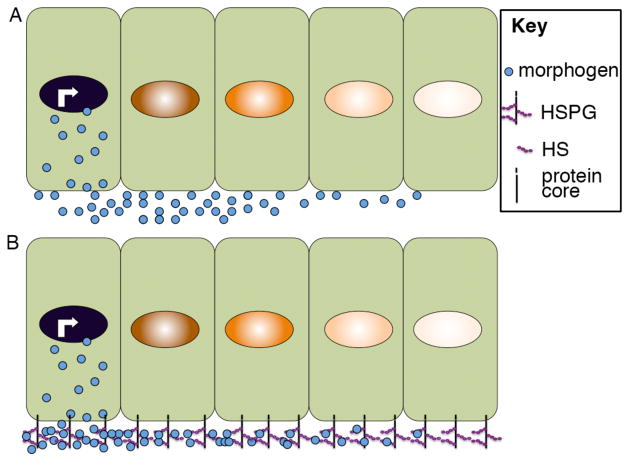
Figure 3. Extracellular movement of morphogens assisted by interactions with HSPGs.
Illustration of morphogen movement through the extracellular space by free diffusion (A) or facilitated by interactions with HSPGs (B). Illustration is schematic and not meant to imply that morphogen movement is restricted to one side of the cell.
Transport facilitated by interactions with HSPGs
Genetic and cell biological studies have shown that heparan sulfate proteoglycans (HSPGs) play an essential role in extracellular movement of many developmental signals including Hh, Wnt/Wg, FGF and bone morphogenetic proteins [Bmps or Decapentaplegic (Dpp) in flies]37. Assessment of the specific role of HSPGs in ligand transport is complicated by the fact that these proteins may also be required for signal transduction, and can affect morphogen stability, endocytosis and intracellular trafficking. This section provides a brief overview of what is known about the role of HSPGS in promoting restricted diffusion. A more comprehensive review of this topic can be found in a recent article by Yan and Lin38.
HSPGs consist of a protein core to which heparan sulfate (HS) and and other glycosaminoglycan (GAG) chains are attached (Fig. 3B). Three major classes of HSPGs have been described-glypicans, syndecans and perlecans. Glypicans are tethered to the cell membrane by a glycophosphatidylinositol (GPI) anchor while syndecans are single-pass transmembrane domain proteins and perlecans are secreted proteins that are deposited into the extracellular matrix. Among these, glypicans play a major role in regulating gradient formation. This function has been best studied in the Drosophila wing disc, where it is possible to directly image the movement of morphogens across a simple field of epithelial cells. The Drosophila glypicans Division abnormally delayed (Dally) and Dally-like (Dlp) are believed to be essential for movement of Hh, Wg and Dpp in the wing disc, as evidenced by the fact that these morphogens are unable to move across the surface of clones of cells mutant for dally and/or dlp39–41. HS modifications are clearly important for this function since mutations in GAG biosynthetic enzymes, or mutations in ligands that abolish their ability to bind to GAG chains lead to defective morphogen signaling and gradient formation in both vertebrates and invertebrates38, 42. While a role for HSPGs in promoting morphogen movement has been well documented in the context of the Drosophila wing disc, and in some cases in vertebrates42 the mechanism by which they do so is less well understood. One possibility is that the morphogen is simply moving down its concentration gradient by being passively displaced from GAG chains on cells near the source and then binding to GAG chains on more distal cells (Figure 3B). Alternatively, it is conceivable that the GPI linkage on glypicans might allow for jumping of HSPG-bound ligand from cell to cell since GPI-linked proteins can be released from the cell surface by the action of phospholipases, or can be transferred intact between the plasma membrane of adjacent cells43. Other classes of membrane anchored HSPGs may function non-cell autonomously to transfer ligands to receiving cells in a similar fashion. In early mouse embryos, for example, cell surface-tethered HS chains are essential for local reception of FGF signals and can also transmit FGF signals to nearby cells in a fashion that is dependent on serine protease activity44. Finally, HSPGs may function more generally to promote the stability of morphogens, affording them more time to diffuse away from producing cells prior to being degraded. Consistent with this last possibility, HSPGs stabilize Wnts in the extracellular space by preventing aggregation45 and Dally has been suggested to compete with and protect Dpp from receptor mediated endocytosis and degradation46. Accessory proteins that enhance ligand stability and diffusion by modulating their interactions with HSPGs have also been identified. The secreted protein shifted, for example, binds to and promotes the stability and signalling range of Hh in the Drosophila wing disc, perhaps by strengthening its interactions with HSPGs47, 48. Another secreted protein, Pentagon, interacts directly with Dally and this leads to enhanced stability and spread of Dpp protein away from its source49. The situation may be more complex in some cases, however, since binding to HSPGs has also been shown to accelerate degradation and to restrict movement of ligands. For example, the mammalian ortholog of Dlp, Glypican-3 (GPC3), competes with the Sonic hedgehog (Shh) receptor, patched, for ligand binding and targets Shh for endocytosis and degradation50. Similarly, interactions with HSPGs enhance internalization and degradation of vertebrate Bmps51 whereas deletion of the HSPG binding motif in Bmp4 generates a ligand with an expanded range of action in both frogs and mice52, 53. Enzymatic digestion of HS chains leads to a similar expansion of the range of FGF movement in zebrafish embryos54. Further studies will be required to determine whether these seemingly opposite effects of HSPGs in either promoting or inhibiting long range movement of morphogens reflect tissue- or species-specific functions or whether they can be accounted for by differences in experimental design.
Facilitated transport of lipid-linked morphogens
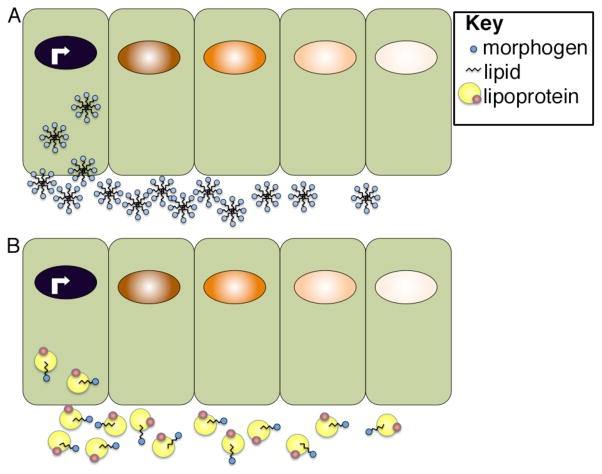
Lipid containing vehicles are likely to participate in transporting a subset of morphogens, such as Hh and Wnts, that are post-translationally modified by addition the addition of palmitic acid and/or cholesterol55, 56. These modifications have been shown to be essential for morphogen movement and for signaling and yet they present a problem in that these lipid-modified proteins must acquire a soluble form in order to travel by diffusion. One mechanism by which they have been proposed to become water-soluble is by assembling into multimeric aggregrates with the hydrophobic lipid moiety positioned on the inside and hydrophilic residues on the outside57–59 (Fig. 4A). Another proposed solution to the problem of insolubility is to package lipid-linked proteins inside of vesicles so that they are surrounded by a membrane bilayer. These export vesicles, called exosomes, could be formed when vesicles carrying newly synthesized lipid-linked proteins from the golgi become incoporated into multivesicular bodies (MVBs). The cargo-carrying vesicles would then be released during exocytosis, when MVBs fuse with the plasma membrane. In the Drosophila wing disc, Wg is detected inside of vesicles that were originally proposed to be specialized exosomes named argosomes60. This model has been updated as more recent studies have shown that these vesicular structures are instead lipoprotein complexes that may function to transport both Wg and Hh (Fig. 4B)61. Lipoproteins are large macromolecules consisting of a central core of hydrophobic lipids surrounded by an outer layer of phospholipids, cholesterol and held together by specialized proteins called apolipoproteins. Lipoprotein particles are best known for their role in transporting cholesterol and other lipids from their site of synthesis in the liver, intestine or fat body to different target sites in the body via the blood stream. Once they reach the target tissue, the lipid cargo is delivered via lipoprotein-receptor uptake and is used in membrane formation, steroid hormone biosynthesis, or post-translational modification of lipidated proteins. In Drosophila, Wnt and Hh proteins have been shown to co-fractionate with lipoprotein particles in the wing disc and reductions in lipoprotein levels causes a loss of long-range but not short-range signaling, suggesting lipoproteins are required for carrying these morphogens outside of cells61, 62. Definitive evidence that lipoproteins function as intercellular carriers of lipidated morphogens in vertebrate organisms is limited, although proteomic analysis has shown that Indian hedgehog co-fractionates with very low density lipoprotein in human plasma63. Furthermore, in early mouse embryos Shh has been detected outside of cells in lipid containing particles that resemble lipoproteins64. Interestingly, in Drosophila wing discs Hh containing lipoprotein particles are recruited to the membrane by binding to HS moieties on Dally and these Hh containing particles remain bound to Dally even after it is shed from the cell surface by cleavage of its GPI anchor. These findings suggested the possibility that HSPGs and lipoproteins might function in concert to transport morphogens outside of cells. This does not appear to be the case, however since the interaction of lipoproteins with Dally does not appear to impact Dally function in facilitating the spread of Hh across the wing disc, although it does enhance Hh signaling65.
Figure 4. Proposed modes of extracellular transport of lipid-linked morphogens.
Lipid-linked morphogens may form micellar-like aggregates in which lipids are positioned on the inside and are surrounded by hydrophobic residues (A) or they may be transported on lipoprotein complexes (B). Illustration is schematic and not meant to imply that morphogen movement is restricted to one side of the cell.
Transport facilitated by soluble binding partners
Many morphogens associate with secreted binding partners that enhance or, more commonly, inhibit their activity and in select cases these proteins have been proposed to serve a dual function as transport vehicles. Dpp, for example, is expressed in dorsal and lateral regions of early Drosophila embryos while the gene encoding the Dpp binding protein Short gastrulation (Sog), is expressed in adjacent ventrolateral cells (Fig. 5). Sog plays both positive and negative roles in regulating Dpp activity. First, it binds to Dpp and blocks its interaction with receptors and prevents the signal from spreading toward ventral cells. At the same time, it facilitates Dpp movement toward dorsal cells. Dpp is released from the inactive complex through the action of the protease tolloid (Fig. 5, yellow lightning bolt), which cleaves Sog 66. As a result of these interactions, peak levels of Dpp ligand are concentrated in a narrow dorsal domain. A refined version of this model that includes a third binding protein and preferential transport of heterodimeric ligands, has also been proposed67. These BMP binding partners are conserved in vertebrates and may fulfill a similar transport function in some contexts.
Figure 5. Extracellular transport of Dpp facilitated by Sog.

Sog competes with receptors for binding to Dpp and restricts its ability to signal to ventral cells while facillitating its diffusion toward the dorsal side of the embryo. Tollid cleaves Sog, releasing Dpp and enabling it to bind and activate receptors. Illustration is schematic and not meant to imply that morphogen movement is restricted to one side of the cell.
The primary role of Sog, and its vertebrate ortholog Chordin, is to inhibit the ability of Dpp/Bmp to bind and activate its receptor68. In many tissues, negative feedback loops function to establish mutually exclusive domains of bmp/dpp and chordin/sog transcription on opposite sides of a field of cells (Fig. 5B). This serves to generate overlapping gradients of the ligand and its inhibitor in the extracellular space, which serves to fine-tune the Bmp/Dpp activity gradient69. In certain cases, as described above, the inhibitor also sharpens the gradient by facilitating ligand transport. Chordin and Sog also bind to cell surface HSPGs and this potentiates their ability to antagonize Bmp/Dpp signaling66. It is not known whether or how interactions of Chordin/Sog with HSPGs affect morphogen movement on the cell surface.
Trancytosis
Another mechanism by which morphogens might establish an extracellular gradient is through consecutive rounds of endocytosis into receiving cells, followed by intracellular trafficking and exocytosis (Fig. 6). This mechanism, termed transcytosis, has the obvious advantage of circumventing problems associated with extracellular movement of “sticky” proteins, such as the lipid-modified morphogens described previously. Initial support for this model came from the observation that ligands can be readily detected inside of signal receiving cells located far from the source70, 71. In some cases, more ligand is seen inside of signal receiving cells than is found in the extracellular space. These findings are difficult to interpret, however, since receptor mediated endocytosis of bound ligands is also known to be required to traffic ligands to the lysosome for degradation and this plays an important role in shaping the gradient. In addition, there is growing awareness that although ligand-bound receptors are initially activated at the plasma membrane, the ligand-receptor complex often must be internalized in order to fully activate cytoplasmic components of the transduction cascade and propagate the signal to the nucleus, as discussed later (see Fig. 8). Thus, one cannot readily determine whether ligand that is detected inside of signal receiving cells far from the source got there as a part of the signal transduction or termination process, or whether it was transported inside of cells as a prelude to intracellular trafficking and exocytosis. Recent studies have shown that Dpp ligand can move across clones of cells lacking the Dpp receptor, suggesting that receptor-meditated transcytosis is not required to transport Dpp across the Wing disc72. These findings do not, however, rule out a role for transcytosis that occurs independent of signalling receptors in gradient formation. The finding that Wg protein can be detected inside cells distal from the source, even in the absence of know signaling receptors, lends support to the concept of transcytosis as a means of gradient formation. Additional support has come from studies in Drosophila wing discs showing that blockade of endocytosis within clones of cells prevents morphogen transport across the clone and thus precludes gradient formation71. The interpretation of this experimental result has been called into question by theoretical predictions showing that blocking endocytosis would lead to an increase in the number of receptors present on the cell surface, and that high affinity interactions with these receptors might trap morphogens and indirectly prevent their transport73. More recent kinetic analysis, however, in which the technique of fluorescence recovery after photobleaching was used to visualize the spread of ligand into cells deficient for endocytosis supports transcytosis as a mechanism to generate a robust and stable gradient, at least in the context of the Drosophila wing disc74. Attempts to detect transcytosis in vertebrate embryos have not been successful to date36, 75. Additional experimental evidence will be required to validate this intriguing possibility.
Figure 6. Model for morphogen transport by transcytosis.
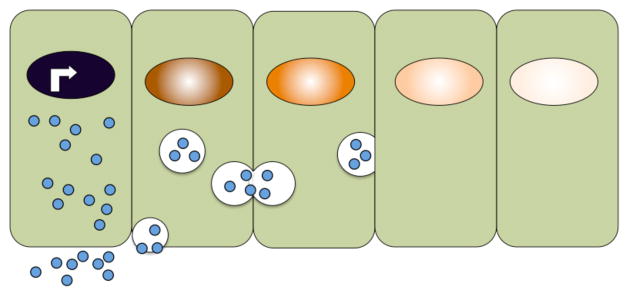
Schematic illustration of a model in which morphogens are actively moved between cells by repeated rounds of secretion, endocytosis and intracellular transport in vesicles.
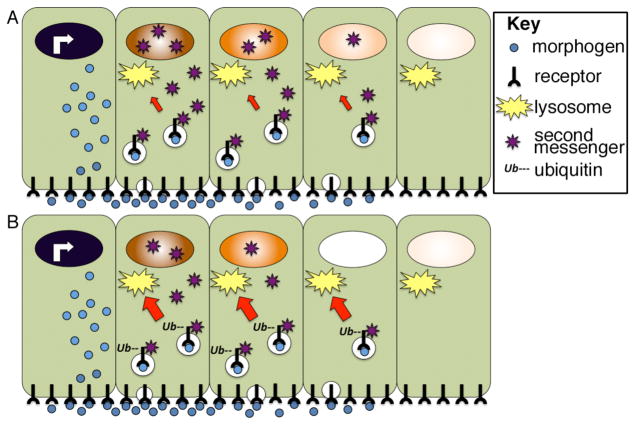
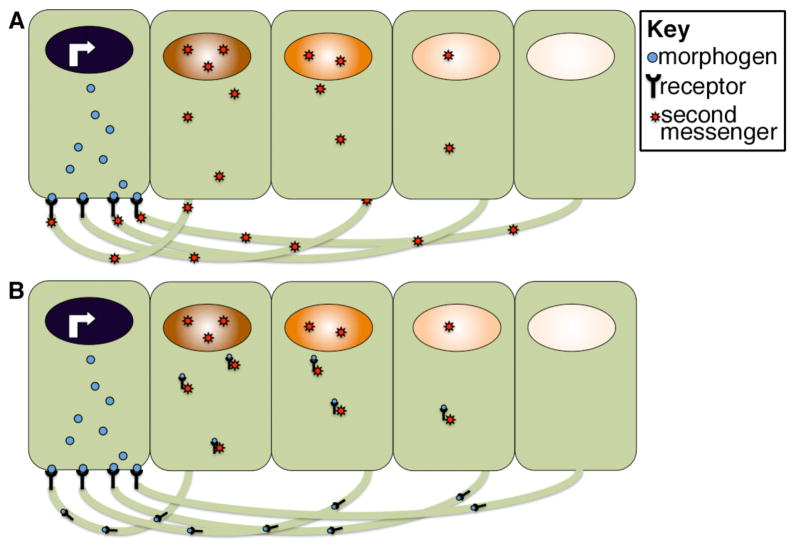
Figure 8. Lysosomal trafficking of activated receptors can shape the morphogen activity gradient.
(A) Following ligand binding, receptor-ligand complexes are internalized and continue to activate second messengers from endosomal compartments. Receptors can be trafficked back to the membrane, or they can be trafficked to the lysosome (red arrow) where they are degraded. (B) Cellular influences that accelerate receptor ubiquitinylation lead to a corresponding increase in the rate of lysosomal trafficking (red arrow) and degradation. This shortens the duration of intracellular signaling, and hence the strength of the nuclear response to an identical extracellular morphogen gradient. Illustration is schematic and not meant to imply that morphogen movement is restricted to one side of the cell.
Cytonemes
A more specialized way in which morphogen gradients may form is through the involvement of structures termed cytonemes. Cytonemes are thin actin-based filopodial bridges that extend across multiple cells to bring membranes from non-adjacent cells into close apposition. Cytonemes were first identified and have been best studied in the Drosophila wing disc where they were hypothesized to be responsible for distributing morphogens76. Specifically, it was hypothesized that morphogens are released from signal sending cells at the site of cytoneme contact and then bind to receptors located on the filapodial membrane to active a second messenger that travels the length of the cytoneme to reach the body of distally located target cells (Fig. 7A). The strength of the signal decays as a function of the distance traveled, generating a gradient of activity in the bodies of target cells. Alternatively, it has been proposed that the ligand-bound receptor might be trafficked back to the cell body to directly activate its signaling cascade in the target cell (Fig. 7B). Consistent with this idea, morphogen receptors have been detected in puncta trafficking along the length of cytonemes where they might conceivably be traveling to the tip to be activated by ligand on signal sending cells, or they might be ferrying the bound ligand back to the target cell77. Similar actin-based filopodia have been detected traversing the blastocoele of early mammalian embryos where they provide a physical connection between cells of the inner cell mass and the trophectoderm78. Morphogen receptors have been immunolocalized to these cell extensions consistent with the possibility that they function to transport or receive signals from ligands released by distal cells as has been proposed in Drosophila78. Cytonemes have the intriguing property of orienting and growing toward morphogen producing cells, suggesting that the morphogen might be functioning as a type of chemoattractant76. Recent work in Drosophila has shown that cells can make distinct subtypes of cytonemes that respond to a specific ligand by segregating the receptor for that particular ligand on the surface of the cytoneme as it grows toward the source79. This raises the interesting possibility that individual cells within a field might simultaneously respond to multiple morphogens by sending out distinct filapodial sensors in several directions. The existence of cytonemes has been documented in a number of organisms, but how or whether they function to sense or distribute morphogens is less clear.
Figure 7. Model for cytoneme function in morphogen gradient formation.
Morphogen receiving cells extend long actin-based filopodia toward morphogen secreting cells. Receptors are proposed to bind the morphogen at points of contact and to either activate second messengers that then traffic back to the body of the target cell (A) or to directly traffic the bound morphogen to receiving cells (B). Illustration is schematic and not meant to imply that morphogen movement is restricted to one side of the cell.
INTERPRETING MORPHOGEN GRADIENTS
Establishing an extracellular ligand gradient is only the first step in assigning positional information to cells within a field. Once the gradient has formed, cells must distinguish small changes in extracellular morphogen concentration, store this information even after the gradient has dissipated, and ultimately transduce this information into qualitatively distinct gene responses inside the nucleus. How this happens is not completely understood but general principals are described below.
In many cases, morphogen concentration outside of cells is directly proportional to differences in the activity of one or a few downstream transcription factors that then regulate distinct target genes in a dose dependent manner. Analysis of how an activin morphogen gradient is interpreted in Xenopus embryos provides a straightforward example of how this might happen80, 81. Canonical activin signals are transduced via a linear membrane to nucleus pathway. Specifically, the ligand binds to and activates a receptor complex that then directly phosphorylates the transcription factor, Smad2, inducing it to form a complex with Smad4. This complex then translocates to the nucleus and activates gene transcription. Individual cells have been shown to perceive activin concentration by assessing the absolute number of occupied receptors on the cell surface. The number of activated receptors determines the relative rate at which Smad2 is phosphorylated, which in turn establishes the Smad2 concentration in the nucleus of a given cell. Thus, in this case activin concentration appears to be translated into a proportional increase in the level of a single activated transcription factor. Notably, many ligand-receptor combinations can activate multiple intracellular signal transduction cascades. In this case, ligand dose could theoretically determine which branch of the cascade is activated to provide qualitative differences in nuclear response, rather than dictating levels of a single activated transcription factor. In general this seems to be the exception rather than the rule.
Extracellular gradients dissipate over time and yet cells cells continue to sense morphogen concentration and remember their position in a gradient even after the ligand is no longer present outside of cells81, 82. The ability of cells to memorize morphogen concentration can be partially accounted for by the fact that most ligand-bound receptors are internalized into the cell and continue to signal from endosomal compartments even after the ligand is removed from the extracellular space83 (Fig. 8A). The duration of this intracellular signaling event can vary and is determined by the rate at which the receptor complex is transported from endosomes (where it can signal) to lysosomes (where it is degraded and can no longer signal). Receptors are marked for lysosomal trafficking by the covalent attachment of ubiquitin residues to their cytoplasmic tails in a reaction catalyzed by an enzyme complex that includes a substrate specific ubiquitin ligase. The relative activity of the ubiquitin ligase determines how long an activated receptor continues to signal inside of the cell and thus significantly impacts the final cellular output in response to a given concentration of ligand83, 84. Various signaling pathways can modify the activity of these enzymes, suggesting a potential mechanism by which different cells could respond differently to the same morphogen concentration due to cross talk between distinct signal transduction cascades85. Specifically, if the ubiquitin ligase that targets a given receptor is upregulated, this would shorten the window during which that receptor could activate downstream second messengers and thereby dampen the nuclear response (Fig. 8B). Whether this mechanism is used to fine tune gradients in vivo, and if so, how widely applicable it is remains speculative.
Morphogen concentration generally dictates the level of activity of a downstream transcriptional effector, but this still leaves open the question of how genes respond to different concentrations of a given transcription factor. There is not one answer to this complicated question. Multiple mechanisms are used to convert transcription factor dosage into distinct gene responses downstream of a given morphogen as described in a comprehensive review by Ashe and Briscoe86. One simple strategy that is employed to elicit distinct gene responses to the same transcription factor exploits differences in the binding affinity of DNA sequences within promoter elements of target genes. In principle, low affinity binding sites will be occupied only in cells where high concentrations of morphogen generate high levels of activation transcription factor whereas high affinity binding sites will be occupied throughout the gradient. A second general strategy is to integrate positive and negative inputs provided by morphogen-triggered transcription factor activation with those from other tissue-specific transcription factors expressed in only a subset of cells. A third strategy relies on secondary interactions between the protein products of the first wave of transcriptional targets. Together, these types of strategies can generate intricate positive and negative feedback circuits and/or secondary overlapping gradients of transcriptional activators and repressors that act on the same set of target genes.
Conclusion
Over the past fifty years, morphogen gradients have evolved from theory to reality. Visualization of the nuclear bicoid gradient provided the first in vivo evidence that morphogens can function as spatial regulators of development8. Most putative morphogens, however, are secreted proteins rather than nuclear factors. In the past decade, elegant studies in Drosophila have directly visualized gradient formation by ectopically expressed, tagged proteins and in some cases by endogenous proteins. By contrast, efforts to directly visualize extracellular gradients of endogenous morphogens in vertebrate species have been less successful and much of our knowledge of gradient formation in vertebrates relies instead on indirect readouts. The identification of specific target genes that are activated downstream of specific ligands, for example, has made it possible to visualize putative gradients of secreted proteins indirectly by looking at the end point of the signaling cascade. More recent discoveries of second messengers that are activated by different ligands, and the development of reagents to detect phosphorylated, active forms of these messengers have taken us a step closer to the ligand, allowing for detection of activity gradients across a field of cells. Experimental studies together with more recent computational analyses have provided support for a diverse number of different strategies by which morphogen gradients are formed, but these are based largely on the results of analyses performed in Drosophila. A challenge for the future will be to use advanced imaging techniques and the growing arsenal of genetic tools to not only test these models more rigorously in Drosophila, but to determine how broadly applicable they are across the animal kingdom.
References
- 1.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 3.Wolpert L. Positional information and patterning revisited. J Theor Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 4.Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwank G, Basler K. Regulation of organ growth by morphogen gradients. Cold Spring Harb Perspect Biol. 2010;2:a001669. doi: 10.1101/cshperspect.a001669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ephrussi A, St Johnston D. Seeing is believing: the bicoid morphogen gradient matures. Cell. 2004;116:143–152. doi: 10.1016/s0092-8674(04)00037-6. [DOI] [PubMed] [Google Scholar]
- 7.Driever W, Nusslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 8.Driever W, Nusslein-Volhard C. A gradient of bicoid protein in Drosophila embryos. Cell. 1988;54:83–93. doi: 10.1016/0092-8674(88)90182-1. [DOI] [PubMed] [Google Scholar]
- 9.Weigmann K, Klapper R, Strasser T, Rickert C, Technau G, Jackle H, Janning W, Klambt C. FlyMove--a new way to look at development of Drosophila. Trends Genet. 2003;19:310–311. doi: 10.1016/S0168-9525(03)00050-7. [DOI] [PubMed] [Google Scholar]
- 10.St Johnston D, Driever W, Berleth T, Richstein S, Nusslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development. 1989;107 (Suppl):13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- 11.Driever W. The bicoid morphogen papers (II): account from Wolfgang Driever. Cell. 2004;116:S7–9. doi: 10.1016/s0092-8674(04)00054-6. 2 p following S9. [DOI] [PubMed] [Google Scholar]
- 12.Driever W, Nusslein-Volhard C. The bicoid protein is a positive regulator of hunchback transcription in the early Drosophila embryo. Nature. 1989;337:138–143. doi: 10.1038/337138a0. [DOI] [PubMed] [Google Scholar]
- 13.Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 14.Nusslein-Volhard CN. The bicoid morphogen papers (I): account from CNV. Cell. 2004;116:S1–5. doi: 10.1016/s0092-8674(04)00055-8. 2 p following S9. [DOI] [PubMed] [Google Scholar]
- 15.Gregor T, Bialek W, de Ruyter van Steveninck RR, Tank DW, Wieschaus EF. Diffusion and scaling during early embryonic pattern formation. Proc Natl Acad Sci U S A. 2005;102:18403–18407. doi: 10.1073/pnas.0509483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130:141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimm O, Coppey M, Wieschaus E. Modelling the Bicoid gradient. Development. 2010;137:2253–2264. doi: 10.1242/dev.032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipshitz HD. Follow the mRNA: a new model for Bicoid gradient formation. Nat Rev Mol Cell Biol. 2009;10:509–512. doi: 10.1038/nrm2730. [DOI] [PubMed] [Google Scholar]
- 20.Houchmandzadeh B, Wieschaus E, Leibler S. Establishment of developmental precision and proportions in the early Drosophila embryo. Nature. 2002;415:798–802. doi: 10.1038/415798a. [DOI] [PubMed] [Google Scholar]
- 21.Spirov A, Fahmy K, Schneider M, Frei E, Noll M, Baumgartner S. Formation of the bicoid morphogen gradient: an mRNA gradient dictates the protein gradient. Development. 2009;136:605–614. doi: 10.1242/dev.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weil TT, Parton R, Davis I, Gavis ER. Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr Biol. 2008;18:1055–1061. doi: 10.1016/j.cub.2008.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lustig KD, Kroll KL, Sun EE, Kirschner MW. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 1996;122:4001–4012. doi: 10.1242/dev.122.12.4001. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- 26.Stennard F, Carnac G, Gurdon JB. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 1996;122:4179–4188. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- 27.Stennard F, Zorn AM, Ryan K, Garrett N, Gurdon JB. Differential expression of VegT and Antipodean protein isoforms in Xenopus. Mech Dev. 1999;86:87–98. doi: 10.1016/s0925-4773(99)00119-7. [DOI] [PubMed] [Google Scholar]
- 28.Hyde CE, Old RW. Regulation of the early expression of the Xenopus nodal-related 1 gene, Xnr1. Development. 2000;127:1221–1229. doi: 10.1242/dev.127.6.1221. [DOI] [PubMed] [Google Scholar]
- 29.Kofron M, Demel T, Xanthos J, Lohr J, Sun B, Sive H, Osada S, Wright C, Wylie C, Heasman J. Mesoderm induction in Xenopus is a zygotic event regulated by maternal VegT via TGFbeta growth factors. Development. 1999;126:5759–5770. doi: 10.1242/dev.126.24.5759. [DOI] [PubMed] [Google Scholar]
- 30.Yasuo H, Lemaire P. A two-step model for the fate determination of presumptive endodermal blastomeres in Xenopus embryos. Curr Biol. 1999;9:869–879. doi: 10.1016/s0960-9822(99)80391-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Houston DW, King ML, Payne C, Wylie C, Heasman J. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 1998;94:515–524. doi: 10.1016/s0092-8674(00)81592-5. [DOI] [PubMed] [Google Scholar]
- 32.Dubrulle J, Pourquie O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- 33.Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- 34.Torroja C, Gorfinkiel N, Guerrero I. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development. 2004;131:2395–2408. doi: 10.1242/dev.01102. [DOI] [PubMed] [Google Scholar]
- 35.McDowell N, Gurdon JB, Grainger DJ. Formation of a functional morphogen gradient by a passive process in tissue from the early Xenopus embryo. Int J Dev Biol. 2001;45:199–207. [PubMed] [Google Scholar]
- 36.Williams PH, Hagemann A, Gonzalez-Gaitan M, Smith JC. Visualizing long-range movement of the morphogen Xnr2 in the Xenopus embryo. Curr Biol. 2004;14:1916–1923. doi: 10.1016/j.cub.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 37.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belenkaya TY, Han C, Yan D, Opoka RJ, Khodoun M, Liu H, Lin X. Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell. 2004;119:231–244. doi: 10.1016/j.cell.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 40.Han C, Belenkaya TY, Wang B, Lin X. Drosophila glypicans control the cell-to-cell movement of Hedgehog by a dynamin-independent process. Development. 2004;131:601–611. doi: 10.1242/dev.00958. [DOI] [PubMed] [Google Scholar]
- 41.Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- 42.Marjoram L, Wright C. Rapid differential transport of Nodal and Lefty on sulfated proteoglycan-rich extracellular matrix regulates left-right asymmetry in Xenopus. Development. 2011;138:475–485. doi: 10.1242/dev.056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kooyman DL, Byrne GW, McClellan S, Nielsen D, Tone M, Waldmann H, Coffman TM, McCurry KR, Platt JL, Logan JS. In vivo transfer of GPI-linked complement restriction factors from erythrocytes to the endothelium. Science. 1995;269:89–92. doi: 10.1126/science.7541557. [DOI] [PubMed] [Google Scholar]
- 44.Shimokawa K, Kimura-Yoshida C, Nagai N, Mukai K, Matsubara K, Watanabe H, Matsuda Y, Mochida K, Matsuo I. Cell Surface Heparan Sulfate Chains Regulate Local Reception of FGF Signaling in the Mouse Embryo. Dev Cell. 2011;21:257–272. doi: 10.1016/j.devcel.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 45.Fuerer C, Habib SJ, Nusse R. A study on the interactions between heparan sulfate proteoglycans and Wnt proteins. Dev Dyn. 2010;239:184–190. doi: 10.1002/dvdy.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 2008;313:408–419. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glise B, Miller CA, Crozatier M, Halbisen MA, Wise S, Olson DJ, Vincent A, Blair SS. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Gorfinkiel N, Sierra J, Callejo A, Ibanez C, Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev Cell. 2005;8:241–253. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 49.Vuilleumier R, Springhorn A, Patterson L, Koidl S, Hammerschmidt M, Affolter M, Pyrowolakis G. Control of Dpp morphogen signalling by a secreted feedback regulator. Nat Cell Biol. 2010;12:611–617. doi: 10.1038/ncb2064. [DOI] [PubMed] [Google Scholar]
- 50.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Jiao X, Billings PC, O’Connell MP, Kaplan FS, Shore EM, Glaser DL. Heparan sulfate proteoglycans (HSPGs) modulate BMP2 osteogenic bioactivity in C2C12 cells. J Biol Chem. 2007;282:1080–1086. doi: 10.1074/jbc.M513414200. [DOI] [PubMed] [Google Scholar]
- 52.Hu Q, Ueno N, Behringer RR. Restriction of BMP4 activity domains in the developing neural tube of the mouse embryo. EMBO Rep. 2004;5:734–739. doi: 10.1038/sj.embor.7400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohkawara B, Iemura S, ten Dijke P, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr Biol. 2002;12:205–209. doi: 10.1016/s0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- 54.Yu SR, Burkhardt M, Nowak M, Ries J, Petrasek Z, Scholpp S, Schwille P, Brand M. Fgf8 morphogen gradient forms by a source-sink mechanism with freely diffusing molecules. Nature. 2009;461:533–536. doi: 10.1038/nature08391. [DOI] [PubMed] [Google Scholar]
- 55.Steinhauer J, Treisman JE. Lipid-modified morphogens: functions of fats. Curr Opin Genet Dev. 2009;19:308–314. doi: 10.1016/j.gde.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eaton S. Multiple roles for lipids in the Hedgehog signalling pathway. Nat Rev Mol Cell Biol. 2008;9:437–445. doi: 10.1038/nrm2414. [DOI] [PubMed] [Google Scholar]
- 57.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 2004;18:641–659. doi: 10.1101/gad.1185804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng J, White B, Tyurina OV, Guner B, Larson T, Lee HY, Karlstrom RO, Kohtz JD. Synergistic and antagonistic roles of the Sonic hedgehog N- and C-terminal lipids. Development. 2004;131:4357–4370. doi: 10.1242/dev.01301. [DOI] [PubMed] [Google Scholar]
- 59.Zeng X, Goetz JA, Suber LM, Scott WJ, Jr, Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411:716–720. doi: 10.1038/35079648. [DOI] [PubMed] [Google Scholar]
- 60.Greco V, Hannus M, Eaton S. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell. 2001;106:633–645. doi: 10.1016/s0092-8674(01)00484-6. [DOI] [PubMed] [Google Scholar]
- 61.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 62.Callejo A, Culi J, Guerrero I. Patched, the receptor of Hedgehog, is a lipoprotein receptor. Proc Natl Acad Sci U S A. 2008;105:912–917. doi: 10.1073/pnas.0705603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Queiroz KC, Tio RA, Zeebregts CJ, Bijlsma MF, Zijlstra F, Badlou B, de Vries M, Ferreira CV, Spek CA, Peppelenbosch MP, et al. Human plasma very low density lipoprotein carries Indian hedgehog. J Proteome Res. 2010;9:6052–6059. doi: 10.1021/pr100403q. [DOI] [PubMed] [Google Scholar]
- 64.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 65.Eugster C, Panakova D, Mahmoud A, Eaton S. Lipoprotein-heparan sulfate interactions in the Hh pathway. Dev Cell. 2007;13:57–71. doi: 10.1016/j.devcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 66.Holley SA, Neul JL, Attisano L, Wrana JL, Sasai Y, O’Connor MB, De Robertis EM, Ferguson EL. The Xenopus dorsalizing factor noggin ventralizes Drosophila embryos by preventing DPP from activating its receptor. Cell. 1996;86:607–617. doi: 10.1016/s0092-8674(00)80134-8. [DOI] [PubMed] [Google Scholar]
- 67.Shimmi O, Umulis D, Othmer H, O’Connor MB. Facilitated transport of a Dpp/Scw heterodimer by Sog/Tsg leads to robust patterning of the Drosophila blastoderm embryo. Cell. 2005;120:873–886. doi: 10.1016/j.cell.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.DeRobertis EM, Sasai Y. A common plan for dorsoventral patterning in bilateria. Nature. 1996;380:37–40. doi: 10.1038/380037a0. [DOI] [PubMed] [Google Scholar]
- 69.Plouhinec JL, De Robertis EM. Systems biology of the self-regulating morphogenetic gradient of the Xenopus gastrula. Cold Spring Harb Perspect Biol. 2009;1:a001701. doi: 10.1101/cshperspect.a001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bejsovec A, Wieschaus E. Signaling activities of the Drosophila wingless gene are separately mutable and appear to be transduced at the cell surface. Genetics. 1995;139:309–320. doi: 10.1093/genetics/139.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Entchev EV, Schwabedissen A, Gonzalez-Gaitan M. Gradient formation of the TGF-beta homolog Dpp. Cell. 2000;103:981–991. doi: 10.1016/s0092-8674(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 72.Schwank G, Dalessi S, Yang SF, Yagi R, de Lachapelle AM, Affolter M, Bergmann S, Basler K. Formation of the long range dpp morphogen gradient. PLoS Biol. 2011;9:e1001111. doi: 10.1371/journal.pbio.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lander AD, Nie Q, Wan FY. Do morphogen gradients arise by diffusion? Dev Cell. 2002;2:785–796. doi: 10.1016/s1534-5807(02)00179-x. [DOI] [PubMed] [Google Scholar]
- 74.Kicheva A, Pantazis P, Bollenbach T, Kalaidzidis Y, Bittig T, Julicher F, Gonzalez-Gaitan M. Kinetics of morphogen gradient formation. Science. 2007;315:521–525. doi: 10.1126/science.1135774. [DOI] [PubMed] [Google Scholar]
- 75.Hagemann AI, Xu X, Nentwich O, Hyvonen M, Smith JC. Rab5-mediated endocytosis of activin is not required for gene activation or long-range signalling in Xenopus. Development. 2009;136:2803–2813. doi: 10.1242/dev.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramirez-Weber FA, Kornberg TB. Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell. 1999;97:599–607. doi: 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- 77.Hsiung F, Ramirez-Weber FA, Iwaki DD, Kornberg TB. Dependence of Drosophila wing imaginal disc cytonemes on Decapentaplegic. Nature. 2005;437:560–563. doi: 10.1038/nature03951. [DOI] [PubMed] [Google Scholar]
- 78.Salas-Vidal E, Lomeli H. Imaging filopodia dynamics in the mouse blastocyst. Dev Biol. 2004;265:75–89. doi: 10.1016/j.ydbio.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 79.Roy S, Hsiung F, Kornberg TB. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332:354–358. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bourillot PY, Garrett N, Gurdon JB. A changing morphogen gradient is interpreted by continuous transduction flow. Development. 2002;129:2167–2180. doi: 10.1242/dev.129.9.2167. [DOI] [PubMed] [Google Scholar]
- 81.Dyson S, Gurdon JB. The interpretation of position in a morphogen gradient as revealed by occupancy of activin receptors. Cell. 1998;93:557–568. doi: 10.1016/s0092-8674(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 82.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 83.Jullien J, Gurdon J. Morphogen gradient interpretation by a regulated trafficking step during ligand-receptor transduction. Genes Dev. 2005;19:2682–2694. doi: 10.1101/gad.341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nowak M, Machate A, Yu SR, Gupta M, Brand M. Interpretation of the FGF8 morphogen gradient is regulated by endocytic trafficking. Nat Cell Biol. 2011;13:153–158. doi: 10.1038/ncb2155. [DOI] [PubMed] [Google Scholar]
- 85.Rainero E, Norman JC. New roles for lysosomal trafficking in morphogen gradient sensing. Sci Signal. 2011;4:pe24. doi: 10.1126/scisignal.2002053. [DOI] [PubMed] [Google Scholar]
- 86.Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
Further Reading/Resources
- 1.Gilbert . Developmental Biology. 9. 2010. [Google Scholar]
- 2.The interactive fly. http://www.sdbonline.org/fly/aimain/1aahome.htm.
- 3.Port F, Basler K. Wnt trafficking: new insights into Wnt maturation, secretion and spreading. Traffic. 2010;11:1265–1271. doi: 10.1111/j.1600-0854.2010.01076.x. [DOI] [PubMed] [Google Scholar]
- 4.Gallet A. Hedgehog morphogen: from secretion to reception. Trends Cell Biol. 2011;21:238–246. doi: 10.1016/j.tcb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JC. Forming and interpreting gradients in the early Xenopus embryo. Cold Spring Harb Perspect Biol. 2009;1:a002477. doi: 10.1101/cshperspect.a002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barkai N, Shilo BZ. Robust generation and decoding of morphogen gradients. Cold Spring Harb Perspect Biol. 2009;1:a001990. doi: 10.1101/cshperspect.a001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Constam DB. Running the gauntlet: an overview of the modalities of travel employed by the putative morphogen Nodal. Curr Opin Genet Dev. 2009;19:302–307. doi: 10.1016/j.gde.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Benazet JD, Zeller R. Vertebrate limb development: moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb Perspect Biol. 2009;1:a001339. doi: 10.1101/cshperspect.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]