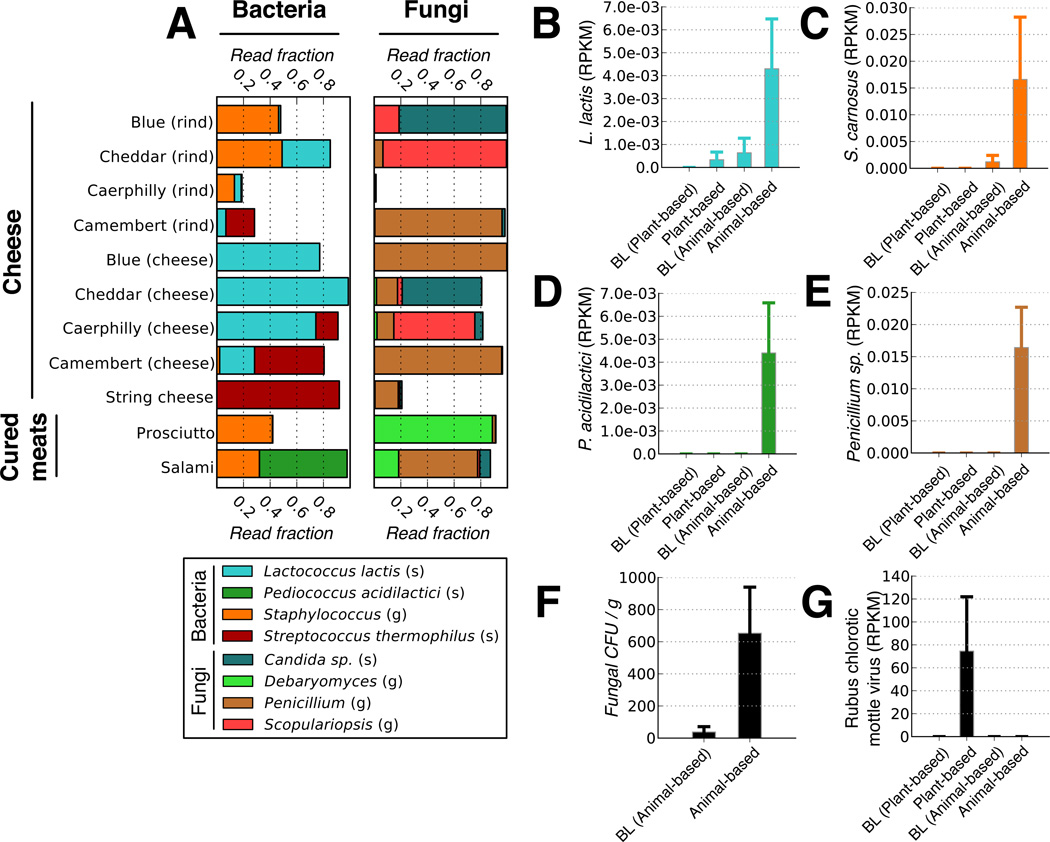
Fig. 4. Foodborne microbes are detectable in the distal gut.
(A) Common bacteria and fungi associated with the animal-based diet menu items, as measured by 16S rRNA and ITS gene sequencing, respectively. Taxa are identified on the genus (g) and species (s) level. A full list of foodborne fungi and bacteria on the animal-based diet can be found in Supplementary Table 21. Foods on the plant-based diet were dominated by matches to the Streptophyta, which derive from chloroplasts within plant matter (Extended Data Fig. 7a). (B-E). Fecal RNA transcripts were significantly enriched (q<0.1, Kruskal-Wallis test; n=6–10 samples/diet arm) for several food-associated microbes on the animal-based diet relative to baseline (BL) periods, including (B) Lactococcus lactis, (C) Staphylococcus carnosus, (D) Pediococcus acidilactici, and (E) a Penicillium sp. A complete table of taxa with significant expression differences can be found in Supplementary Table 22. (F) Fungal concentrations in feces before and 1–2 days after the animal-based diet were also measured using culture media selective for fungal growth (plate count agar with milk, salt, and chloramphenicol). Post-diet fecal samples exhibit significantly higher fungal concentrations than baseline samples (p<0.02; two-sided Mann-Whitney U test; n=7–10 samples/diet arm). (G) Increased RNA transcripts from the plant-derived Rubus chlorotic mottle virus transcripts increase on the plant-based diet (q<0.1, Kruskal-Wallis test; n=6–10 samples/diet arm). Barplots (B-G) all display mean±sem.