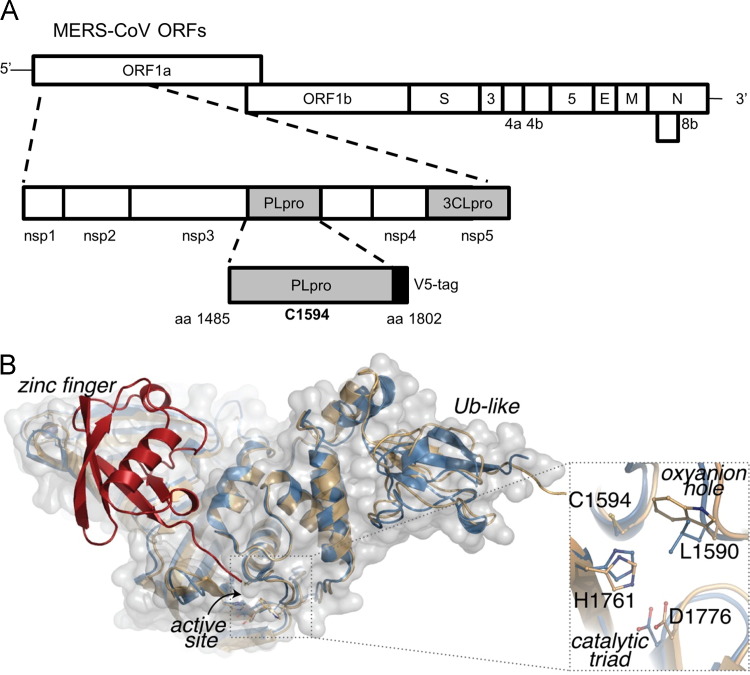
Fig. 1.
Modeling MERS-CoV PLpro onto the SARS-CoV PLpro-ubiquitin-aldehyde structure. (A) Schematic diagram of MERS-CoV ORFs and the papain-like protease (PLpro) domain within nonstructural protein 3 (nsp3). Expression plasmid pcDNA-MERS-PLpro (amino acids 1485–1802) and the predicted catalytic cysteine residue 1594 are indicated. (B) Homology model of MERS-CoV PLpro (blue cartoom and gray surface) aligns with the overall structural architecture found in SARS-CoV PLpro-ubiquitin-aldehyde complex PDB:4MM3 (beige cartoon), including the ubiquitn binding domain at the zinc finger and the extended Ub-like (Ubl) domain. Ubiquitin (red) modeled into the zinc finger domain of MERS-CoV PLpro, with its C-terminus reaching the active site. An enlargement of predicted MERS-CoV PLpro active site superimposed onto the SARS-CoV PLpro active site suggests that the MERS-CoV PLpro catalytic triad is composed of C1594–H1761–D1776 and the putative oxyanion hole residue is L1590.