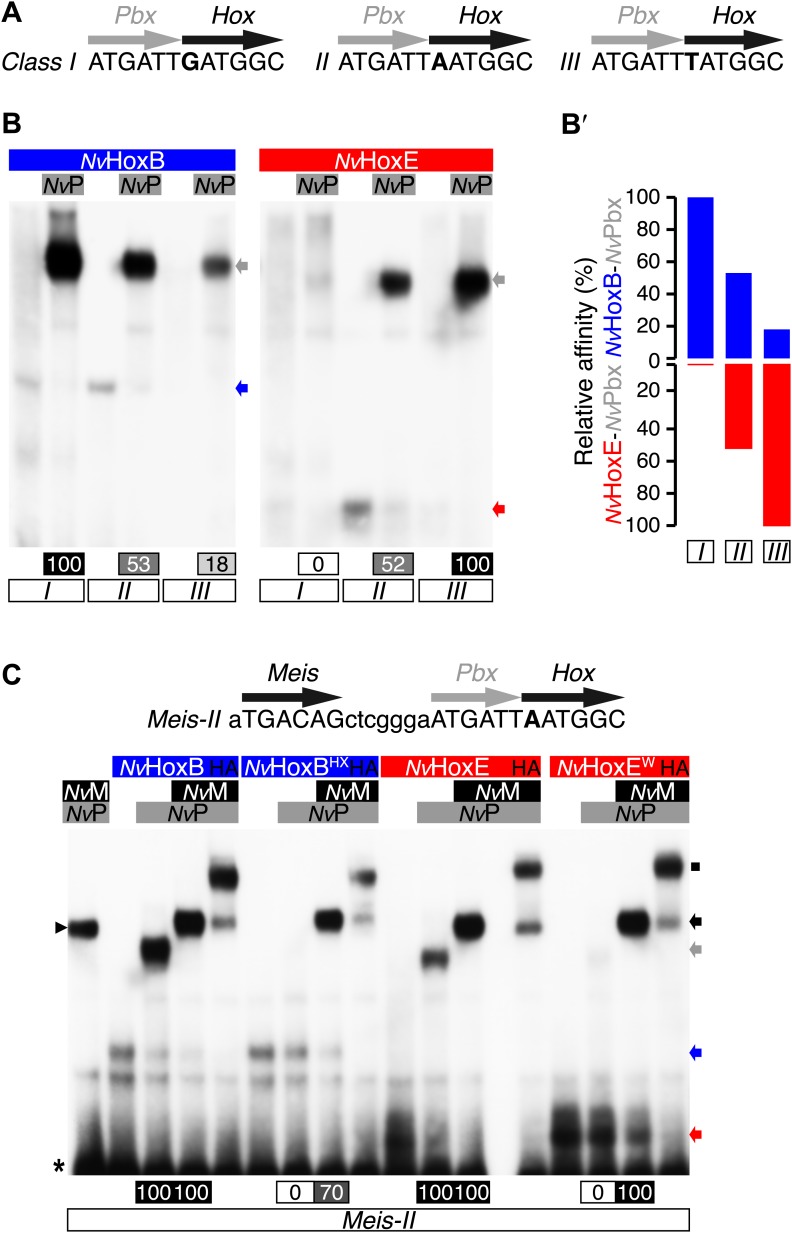
Figure 4. Interaction properties between NvHox and NvTALE proteins in vitro.
(A) Nucleotide sequence of the different classes of Hox/Pbx binding sites used in band shift experiments. The nucleotide that distinguishes each Hox/Pbx binding site is bolded. (B–B′) Band shift experiments between NvHoxB or NvHoxE and NvPbx on the three different classes of binding sites, as indicated. Coloured and grey arrows point to monomer or dimer binding, respectively. Graph on the right (B′) depicts the relative affinity of each dimeric complex on the three different binding sites, as deduced from the direct quantification on the gel (values are indicated at the bottom). (C) Band shift experiments between wild-type or HX-mutated NvHox proteins and NvTALE cofactors, as indicated. Colour codes and annotations are as in (B). Black arrow indicates trimeric NvHox/NvPbx/NvMeis complexes. Other bands are not specific (proteins of the lysate). Black scare highlights the supershift band resulting from the addition of an antibody against the HA tag of NvHox proteins. Asterisk shows the free probe. Note that the loss of dimeric NvHox/NvPbx complex upon the HX mutation is rescued in the presence of NvMeis.