Figure 5. Genes expressed in the endoderm are enriched in Hox-TALE binding sites in their promoter region.
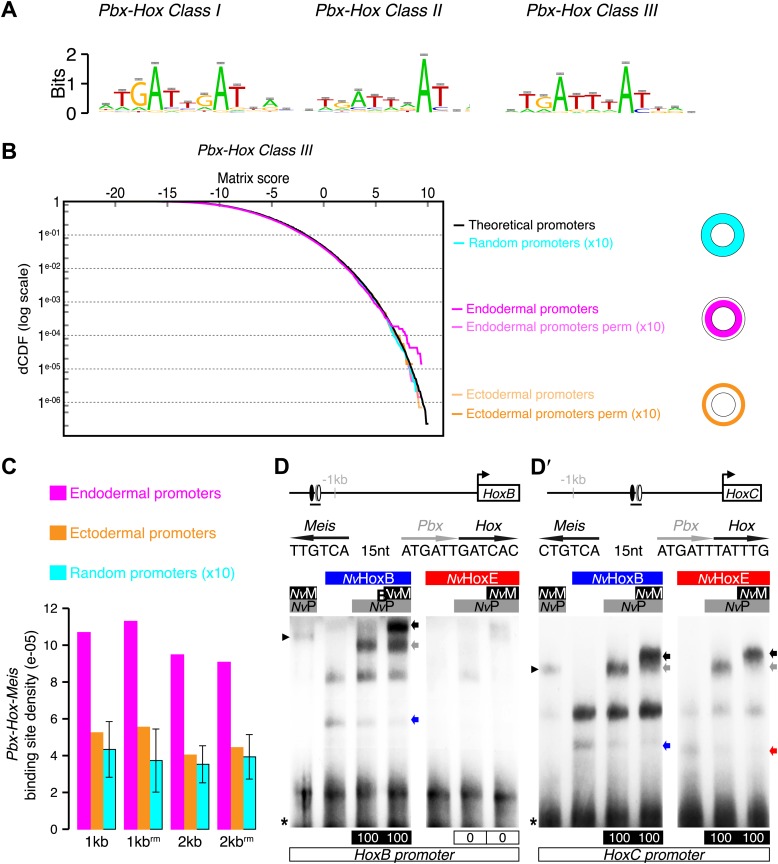
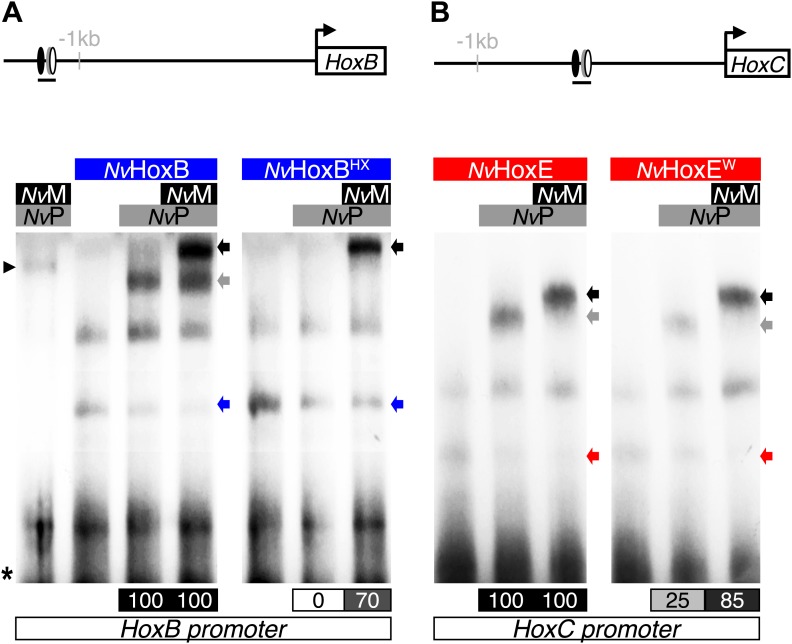
(A) Hox/Pbx binding motifs represented as logos. The three motifs represent the binding specificity of the Hox/Pbx complex for sites of class I, II, or III. Matrix was determined by Selex with the Drosophila proteins (Slattery et al., 2011). (B) Score distributions of the Hox/Pbx Class III matrix. The Y-axis is shown in logarithmic scale to highlight the relevant range of p values (small values). The separation of the pink curve (endoderm) from the black one (theoretical distribution) indicates an enrichment of the Hox/Pbx putative binding sites in the promoter of genes expressed in the endoderm. On the contrary, there is no enrichment in the promoter of genes expressed in the ectoderm, as the orange curve follows the black one. All negative controls also show no enrichment: random sets of gene promoters (cyan), promoter regions randomized by matrix column permutations for the endoderm (light pink) and ectoderm (light orange). (C) In silico analysis of Hox/Pbx/Meis binding sites in the promoter region (1 kb or 2 kbs upstream of the transcription start site) of genes expressed in the endoderm (pink), ectoderm (orange), or randomly chosen (cyan). The graph illustrates the preferential enrichment of Hox/Pbx/Meis binding sites in the promoter region of endodermal genes. Rm: repeat masked. (D–D′) Band shift experiments between NvHox and NvTALE proteins on binding sites found in the promoter region of NvHoxB and NvHoxC genes. Sequence and genomic position of each binding site are shown above the gel. Colour code and annotations are as in Figure 3. Note the distinct DNA-binding preferences of NvHoxB and NvHoxE on these two different target sites. See also Figure 5—figure supplements 1 and 2.