Figure 6. Functional analysis of NvHox and NvPbx proteins in Drosophila.
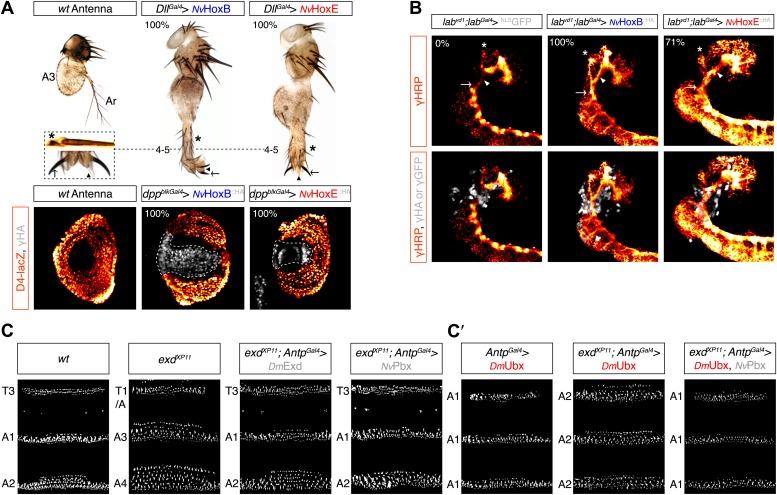
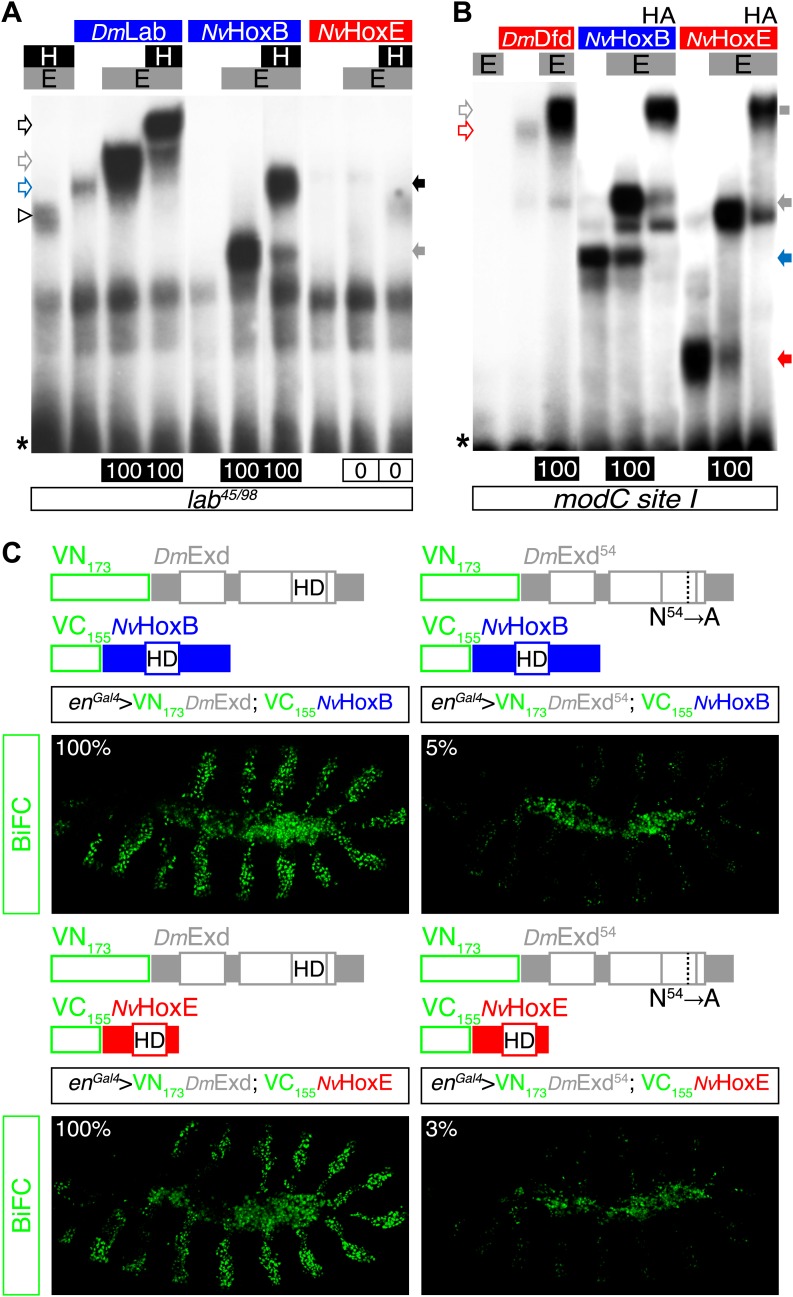
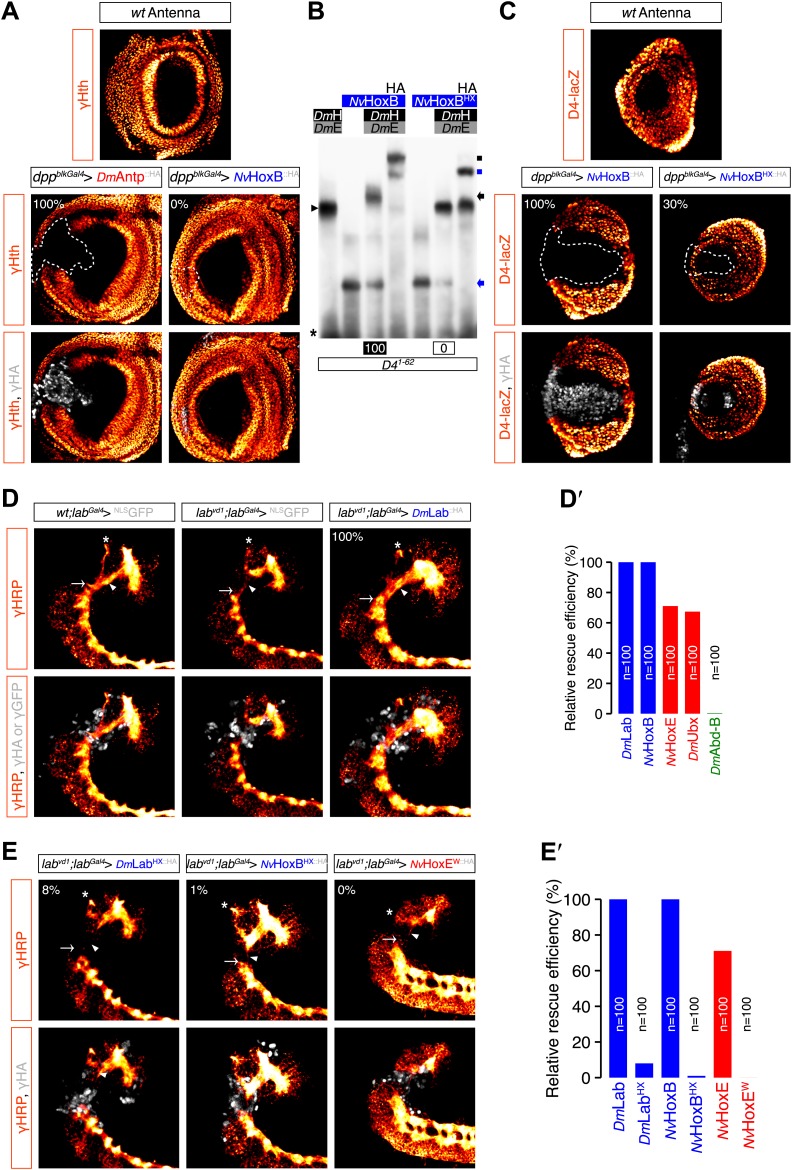
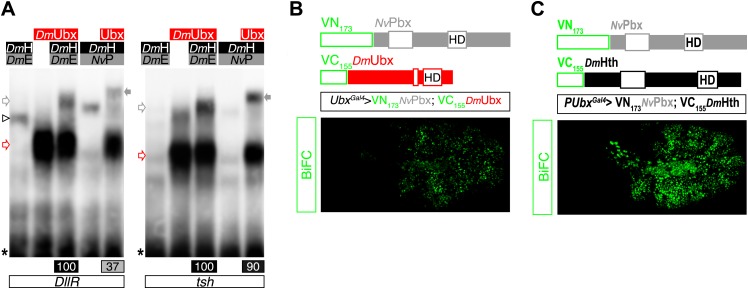
(A) Antenna-to-leg transforming activities of NvHoxB and NvHoxE. NvHox proteins were expressed in the antenna with the Distalless (Dll)-Gal4 driver. Asterisk depicts leg-specific bracted bristles. 4–5 shows the transformation of the arista in two tarsal segments. Arrow and arrowhead in the enlargement indicate the formation of the leg-specific terminal claw and its associated sensory pad respectively. The antenna-to-leg transformation by NvHox proteins (grey) is achieved through the repression of the spineless (ss) target gene, as observed by the repression of the ss enhancer D4 activity on lacZ reporter gene expression (orange). See also Figure 6—figure supplements 1 and 2. (B) Rescue of the labial (lab) mutant phenotype in the tritocerebrum by NvHox proteins. The central nervous system is stained with an anti-HRP (orange). Hox or GFP (as a control) proteins (grey) are expressed in the tritocerebrum with a lab-Gal4 driver. Frontal connectives (asterisk), longitudinal connectives (arrowhead) and tritocerebral commissure (arrow) are indicated. In lab mutant background, longitudinal connectives are reduced, frontal connectives project ectopically and the tritocerebral commissure is missing (Hirth et al., 1998). Expression of NvHoxB or NvHoxE in this mutant context leads to a complete or strong rescue of this phenotype, respectively. See also Figure 6—figure supplement 2. (C–C′) NvPbx can rescue zygotic exd mutant phenotypes in the Drosophila larva cuticle. (C) Larvae homozygous for the zygotic exdXP11 mutation have T3 and A1 segments that resemble to a T1/abdominal or A3 segment, respectively. Thoracic expression of either DmExd or NvPbx in this mutant background (through the UAS/Gal4 system, with the Antennapedia (Antp)-Gal4 driver) is sufficient to restore the correct specification of T3 and A1, as assessed by the shape and arrangement of denticle belts. (C′) Ubx normally specifies the A1 segment. Ectopic expression of Ubx with Antp-Gal4 induces A1-like segments in the thorax. In absence of Exd, Ubx produces A2-like segments. Providing back NvPbx in this genetic background is sufficient to restore the normal A1-inducing activity of Ubx. See also Figure 6—figure supplements 3 and 4.