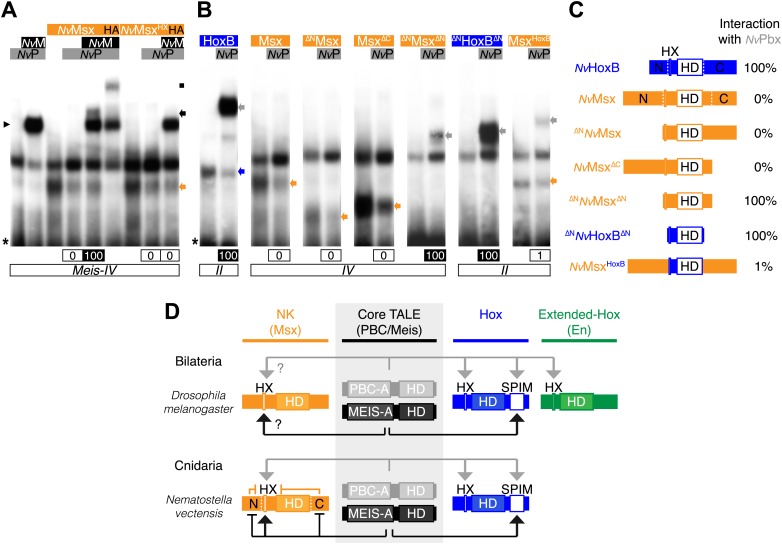
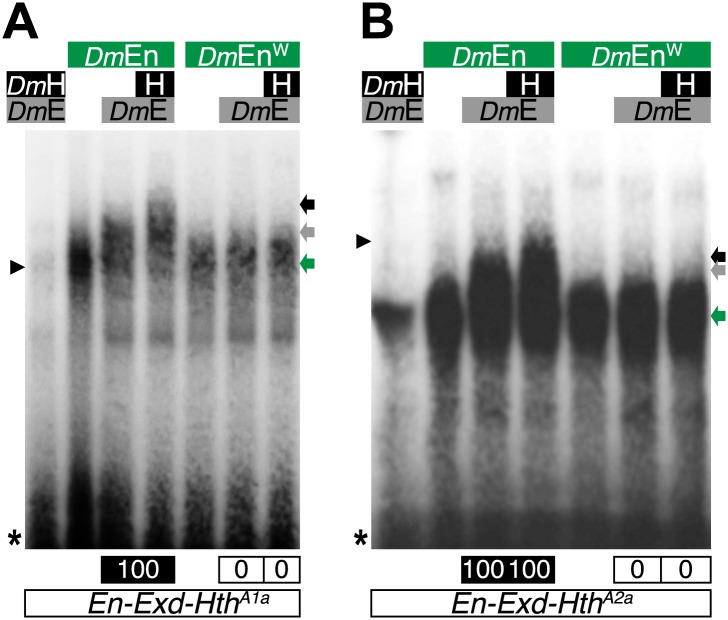
Figure 8. Genesis of Hox–TALE interaction networks in Metazoa.
(A) Band shift experiments between NvMsx and NvTALE cofactors, as indicated. Colour code and annotations are as in previous figures. Note that no dimeric NvMsx/NvPbx complex is formed. Binding reactions with NvMsx proteins were performed on a consensus Msx/Pbx binding site derived from vertebrates and containing an additional Meis binding site in a topology similar to the Hox probe (‘Materials and methods’). (B) Band shift experiments between wild-type, truncated or chimeric NvMsx and/or NvHoxB proteins, and NvPbx, as indicated. (C) Scheme of the diverse protein constructs and their corresponding interaction affinity level with NvPbx, as assessed from quantification of each band shift. Quantifications with truncated NvMsx proteins were deduced by comparison with the trimeric NvMsx/NvPbx/NvMeis in (A). (D) Molecular rules underlying interaction properties between NK, Hox, or extended-Hox members and TALE cofactors in Eumetazoans. In this model, Meis is able to promote HX-dependent interactions between NvMsx and NvPbx by masking inhibitory interaction domains in NvMsx. Whether a similar role could exist in Bilateria remains to be determined. We noticed that the Drosophila Msx protein contains an HX and forms trimeric but not dimeric complexes with the Drosophila TALE partners (not shown). HX-dependency in those interactions remains to be determined (question marks). Interaction between DmEn and PBC is also HX-dependent but does not require the presence of Meis to occur. In contrast to the NK or extended-Hox families, most members of the Hox family have retained a HX motif. This motif is required for generic Hox/Pbx functions. The additional presence of Meis allows revealing specific Pbx interaction motifs (SPIMs), which could be important for distinguishing and/or diversifying the embryonic activities of each Hox paralog group member. See also Figure 8—figure supplement 1.