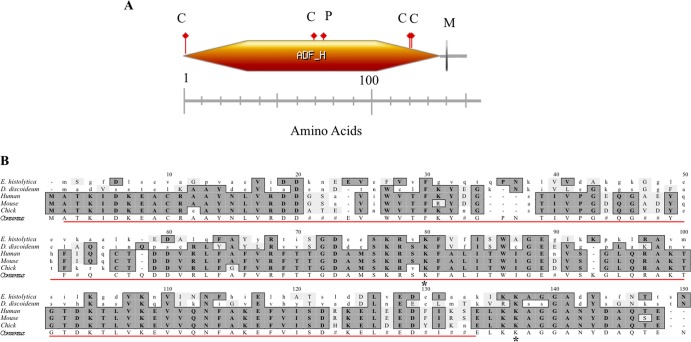
FIG 2.
Domains found in EhCoactosin and sequence comparison of coactosin from E. histolytica to those of other species. (A) EhCoactosin is an F-actin binding protein with a postulated molecular mass of 16.2 kDa (148 amino acids). It is predicted to have an actin depolymerization factor homology (ADF-H) domain (orange hexagon), an N-myristoylation site (gray vertical line labeled M), 4 casein kinase II phosphorylation sites (red flags labeled C), and a protein kinase C phosphorylation site (red flag labeled P). (B) The predicted amino acid sequence of EhCoactosin was aligned with coactosin from other organisms using ClustalW multiple sequence alignment, v1.4 (MacVector v9.0). Conserved amino acids are shaded, and the consensus sequence is indicated below the aligned sequence (#, conservation but no consensus). The E. histolytica ADF-H domain is indicated by the red line. Asterisks represent the lysine residues Lys75 and Lys131 of human coactosin that are essential for binding to F-actin and 5-lipoxygenase, respectively.