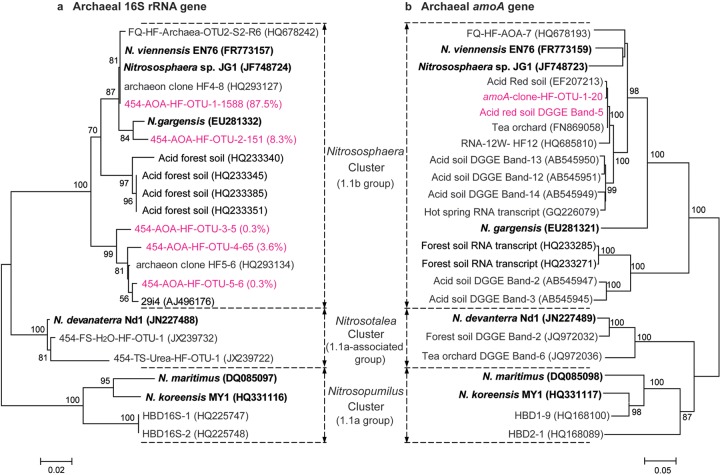
FIG 4.
Phylogenetic analysis of archaeal 16S rRNA (a) and amoA genes (b) in the [13C]DNA from the labeled microcosms after incubation for 56 days. The 16S rRNA reads of archaeal 16S rRNA genes in “heavy” DNA fractions 5, 6, and 7 were pooled for analysis with a total of 1,815 reads. The notation “454-AOA-HF-OTU-1-1588-(87.5%)” indicates that OTU-1 contains 1,588 sequences with >97% identity, accounting for 87.5% of the total archaeal 16S rRNA gene reads retrieved from the [13C]DNA. The notation “amoA-clone-OTU-1-20” indicates that all 20 cloned archaeal amoA genes in the [13C]DNA showed sequence identity of >97% and could be taxonomically classified into a single phylotype. One representative sequence was extracted using the mothur software for the tree construction of the archaeal amoA and 16S rRNA genes. The notation “Acid red soil DGGE band-5” represents the sequence of the newly emerging DGGE bands in the acidic soil studied after incubation for 56 days (data not shown). The putatively active amoA genes within the Nitrososphaera cluster were retrieved from various publications for analysis, including FQ-HF-AOA-7 (22) and RNA-12W-HF-12 (30) in alkaline soil, hot spring RNA transcripts (31), putatively active AOA DGGE bands 12, 13, and 14 in acidic soils (11), and acidic forest soil RNA transcripts (14).