Abstract
White-nose syndrome is a fungal disease that has decimated bat populations across eastern North America. Identification of the etiologic agent, Pseudogymnoascus destructans (formerly Geomyces destructans), in environmental samples is essential to proposed management plans. A major challenge is the presence of closely related species, which are ubiquitous in many soils and cave sediments and often present in high abundance. We present a dual-probe real-time quantitative PCR assay capable of detecting and differentiating P. destructans from closely related fungi in environmental samples from North America. The assay, based on a single nucleotide polymorphism (SNP) specific to P. destructans, is capable of rapid low-level detection from various sampling media, including sediment, fecal samples, wing biopsy specimens, and skin swabs. This method is a highly sensitive, high-throughput method for identifying P. destructans, other Pseudogymnoascus spp., and Geomyces spp. in the environment, providing a fundamental component of research and risk assessment for addressing this disease, as well as other ecological and mycological work on related fungi.
INTRODUCTION
White-nose syndrome (WNS) is a fatal disease of hibernating bats first noted as a white growth on the muzzles and wings of bats in caves near Albany, NY, in 2006 (1). Since then, WNS has emerged as a deadly epizootic that has rapidly spread throughout eastern North America, killing millions of bats (2). The psychrophilic fungus Pseudogymnoascus destructans (formerly Geomyces destructans) was demonstrated to be the etiologic agent of WNS (3, 4), and disease is currently diagnosed by histopathology on bat wing or muzzle tissue (5). Unfortunately, histopathology is an invasive procedure performed on dead bats or skin tissue, requiring highly trained specialists and substantial time for sample preparation and the interpretation of results. A rapid, high-throughput quantitative PCR (qPCR) is essential for proposed continental-scale epizootiological research and disease risk assessment.
One such approach is to noninvasively screen for the presence of P. destructans on bats independent of WNS symptoms, affording the flexibility to monitor bat populations before, during, and after an outbreak. Furthermore, P. destructans may be detectable on bats and in their hibernacula in advance of mortality events. In fact, P. destructans should be present on asymptomatic bats during the incubation period of the fungus, which is thought to be at least several months (3). Pseudogymnoascus destructans has already been detected in the sediment of WNS-affected bat hibernacula (6). Sediment, along with other hibernaculum substrates, may play an important role in the transmission of the disease to healthy bats (6). The detection of P. destructans on bats or in their environment is not diagnostic for WNS but would certainly indicate a potential risk of infection. Early detection opens a window of opportunity to establish the risk of disease in a population and take appropriate action.
Methods for the detection of P. destructans began with culture and microscopy (7) but now include various PCR techniques (8–11). PCR-based tests allow for the rapid and sensitive detection of target DNA from a variety of sample source materials (bat skin swabs, tissue biopsy specimens, cave sediment, hibernaculum surface swabs, feces, etc.). Compared to conventional PCR methods, real-time PCR confers increased specificity by means of a sequence-specific probe and improved sensitivity due to a fluorescent marker. PCR-based assays are limited by the quality and quantity of DNA obtained from sample DNA extractions, and care must be taken during assay design and validation to ensure that tests of genetically related organisms do not result in false-positive signals. The ability to distinguish P. destructans from near-relative Pseudogymnoascus and Geomyces species is particularly relevant due to the ubiquity of these closely related fungi in soil and cave sediments (10, 12).
We developed a dual-probe TaqMan qPCR with the capability of detecting DNA from P. destructans and near relatives at minute quantities and differentiating bat-pathogenic P. destructans from allied fungi in samples collected from across North America. The qPCR targets a single nucleotide polymorphism (SNP) in the internal transcribed spacer 1 (ITS1) region of the genome, one allele being specific for P. destructans and the other specific for allied Pseudogymnoascus and Geomyces. Our use of the TaqMan minor groove binder (MGB) quencher increases probe specificity for this target sequence, and the high copy number of ITS1 in fungi (13) provides for greater sensitivity than single-copy target qPCR assays can achieve. We validated the assay with well-characterized controls to determine qPCR sensitivity, specificity, and capability of resolving mixtures of P. destructans DNA and DNA from other Pseudogymnoascus spp. Assay performance was compared to those of two other real-time PCR assays specific for P. destructans: the α-(l)-rhamnosidase (ALR) PCR (9) and the intergenic spacer (IGS) PCR (11). Unlike these other qPCR assays, the ITS1 qPCR can detect other Pseudogymnoascus spp. and Geomyces spp. in addition to P. destructans, providing a suitable technique for microbial community analysis and other applications outside WNS research.
MATERIALS AND METHODS
SNP discovery.
To identify a P. destructans-specific SNP, we utilized 86 Pseudogymnoascus and Geomyces ITS1 sequences from GenBank (accession numbers HM848924 to HM848997 [10], AJ608972, AM901700, AY345347, AY345348, DQ117444, DQ402527, EF434077, EU884921, FJ362279, FJ545236, JQ034511, and JX131373), including seven sequences from P. destructans clade 1 (EU884921, HM848979, HM848976, HM848977, HM848972, HM848975, and HM848978). We aligned these sequences with Sequencher 4.10.1 software (Gene Codes Corporation, Ann Arbor, MI) and visually identified a single nucleotide deletion of C at position 125 of the EU884921 ITS1 sequence, which was unique to all seven P. destructans sequences. The genetic diversity of the 171-bp ITS1 region of these genera ranged from 77 to 100% identity. The P. destructans sequences were nearly identical to each other, with only one sequence possessing one SNP.
TaqMan SNP genotyping assay development.
We designed a custom dual-probe TaqMan qPCR (Life Technologies, Carlsbad, CA), hereafter called the ITS1-qPCR, to target the P. destructans-specific deletion in ITS1. Primers and probes (Table 1) were designed according to the manufacturer's specifications with Primer Express software (Life Technologies). There were no mismatches between the primers and any of the available Pseudogymnoascus or Geomyces sequences. Also, the only sequence mismatch in the probes occurred in the single base deletion distinguishing P. destructans from the other species. The 6-carboxyfluorescein (FAM)-labeled probe (6FAM_Pd-ITS1_Pd) is specific for the P. destructans allele, whereas the VIC-labeled probe (VIC_Pd-ITS1_Psp) is specific for the allele shared by other species of Pseudogymnoascus and Geomyces fungi. The specificity of the VIC-labeled probe for Pseudogymnoascus spp. and Geomyces spp. was confirmed by comparing the probe to ITS sequences presented in the work of Lorch et al. (14) and Minnis and Lindner (12), as well as G. pannorum sequences from GenBank. The qPCR was optimized on a 7500 Fast Real-Time PCR System (Life Technologies, Carlsbad, CA). Each 10-μl PCR mixture contained 1× TaqMan Universal master mix (Life Technologies), 0.025 U/μl of Platinum Taq (Life Technologies), 0.3 μM each primer, 0.160 μM Pseudogymnoascus sp. probe, 0.116 μM P. destructans probe, and up to 4 μl of template DNA. Thermocycler conditions were as follows: 2 min of UNG activation at 50°C and 10 min of hot start at 95°C, followed by 40 cycles of 15 s of denaturation at 95°C and 1 min of annealing at 62°C. Data were analyzed with 7500 software, version 2.0.6 (Life Technologies). The baseline-corrected normalized reporter signal (ΔRn) threshold used to determine cycle threshold (CT) was set manually for both FAM and VIC signals such that the signal curves intercepted this value at 10% of their maximum amplitude (11). Typically, the ΔRn threshold was set to 0.2 for both signals.
TABLE 1.
Primers and probes utilized in the ITS1-qPCRa
| Name | Function | 5′ label | Sequence (5′–3′) |
|---|---|---|---|
| Gd-ITS1-qPCR_F | Forward primer | NA | CTTTGTTTATTACACTTTGTTGCTTT |
| Gd-ITS1-qPCR_R | Reverse primer | NA | CCGTTGTTGAAAGTTTTAACTATTATAT |
| 6FAM_Pd-ITS1-qPCR_Pd | P. destructans-specific probe | FAM | CTTGCCAGAGGACTAA |
| VIC_Pd-ITS1-qPCR_Psp | Pseudogymnoascus sp./Geomyces sp.-specific probe | VIC | CTTGCCAGAGGACCTA |
Both probes include a nonfluorescent quencher and minor groove binder at the 3′ end. The C residue deleted in P. destructans is underlined in the VIC probe sequence. NA, not applicable.
Assay specificity.
A panel of genomic DNA extracted from pure cultures of Pseudogymnoascus and other fungal strains (Table 2) was used to ascertain the specificity of the qPCR. This panel consisted of DNA from 34 isolates of P. destructans, as confirmed by conventional ITS1 PCR and sequencing (8). Of these 34 isolates, 22 were from the WNS-affected region of North America, and 12 were from Europe. The panel also included DNA from 20 strains of Pseudogymnoascus spp., 2 strains of Geomyces auratus (also known as Geomyces pannorum var. pannorum), 3 strains of Leuconeurospora spp. from North America (10, 12), and 52 other strains representing a broad phylogenetic range of fungi. Genomic DNA was extracted and purified with a number of methods, notably the OmniPrep for Fungi kit (G-Biosciences, St. Louis, MO). DNA was quantified with a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA) and diluted to 2 ng/μl in UltraPure DNase/RNase-free distilled water (Life Technologies) prior to being assayed.
TABLE 2.
Fungal genomic DNA used as controls for the validation of the ITS1-qPCRa
| Taxon or sample description | Strain name or GenBank number | Country | Region |
|---|---|---|---|
| Pseudogymnoascus destructans | 20631-21 | USA | NY |
| 20674-9 | USA | VT | |
| 20682-10 | USA | MA | |
| 20693-1 | USA | MA | |
| 22004-1 | USA | CT | |
| 22426-2 | USA | CT | |
| 22429-8 | USA | WV | |
| 22442-2 | USA | NJ | |
| 22469-1 | USA | VA | |
| 22480-1 | USA | NY | |
| 22504-1 | USA | PA | |
| 22884-4W | USA | VT | |
| 22930-2 | USA | TN | |
| 22948-1 | USA | TN | |
| 22949-4 | USA | MD | |
| 22971-3 | CAN | ON | |
| 22972-2W | CAN | ON | |
| 22997-1 | USA | TN | |
| 23414-1W | USA | IN | |
| 23434-1W | USA | IN | |
| 23444-1 | USA | TN | |
| 23455-1 | USA | VA | |
| AnT23022011-1_Mmyo2 | UKR | ||
| AnT23022011-1_Mmyo4 | UKR | ||
| EST 2 | EST | ||
| GU350433 | CHE | Aargau | |
| GU350434 | HUN | Kislod | |
| GU999986 | DEU | Thuringia | |
| HaR06022011-1 | FRA | ||
| LaA16022011C1.2 | FRA | ||
| Mmyo03042010-FrF-BEL-1 | BEL | ||
| Mmyo04032010-YLB-GLE1-1 | FRA | ||
| Mmyo12032009-PaV-PER-1_4 | FRA | ||
| WiB07032010-SIL-1 | POL | ||
| Pseudogymnoascus sp. | 3629 | ||
| 7792 | |||
| 13PA01 | USA | PA | |
| 14PA05 | USA | PA | |
| 15PA10B | |||
| 17WV03 | USA | WV | |
| 23014-1-I2 | |||
| 23014-1-I6 | |||
| 23342-1-I1 | |||
| 24MN10 | USA | MN | |
| 24MN11 | USA | MN | |
| 24MN13 | USA | MN | |
| 24MN18 | USA | MN | |
| 24MN21 | USA | MN | |
| 24MN24 | USA | MN | |
| 24MN32 | USA | MN | |
| 03VT05 | USA | VT | |
| C101 | |||
| NYI3 | |||
| Geomyces auratus | cbs 108.29 | ||
| Geomyces auratus | |||
| Geomyces sp. | 12NJ08 | USA | NJ |
| Leuconeurospora sp. | 01NH01 | ||
| 01NH04 | |||
| 01NH05 | USA | NH | |
| Antrodiella aff. romellii | |||
| Antrodiella sp. | |||
| Armillaria solidipes | |||
| Bjerkandera adusta | DR5022 | ||
| Camarographium carpini | RG134 | ||
| Chaetomium globosum | RG136 | ||
| Coniophora sp. | RG101 | ||
| Coniothyrium carteri | RG139 | ||
| Coniothyrium carteri | RG140 | ||
| Dacrymyces subalpinus | DR5187 | ||
| Fimetariella sp. | |||
| Geastrumia polystigmatis | RG109 | ||
| Gibberella sp. | RG131 | ||
| Heterobasidion annosum | |||
| Heterobasidion annosum | |||
| Heterobasidion annosum | |||
| Hypochnicium cystidiatum | DR5730 | ||
| Metarhizium flavoviride var. Pemphigi | |||
| Mycenoid agaric | LR 9-1 | ||
| Neonectria faginata | RG01 | ||
| Neonectria faginata | RG112 | ||
| Neonectria punicea | AR4167 | ||
| Nigroporus sp. | |||
| Onnia tomentosa | |||
| Onnia tomentosa | |||
| Onnia tomentosa | |||
| Onnia tomentosa | |||
| Onnia tomentosa | |||
| Onnia tomentosa | |||
| Penicillium fellutanum | RG103 | ||
| Phaeoacremonium sp. | RG107 | ||
| Phoma sp. | RG104 | ||
| Phomopsis | |||
| Pithomyces valparadisiacus | RG108 | ||
| Pluteus sp. | |||
| Pluteus sp. | |||
| Pluteus sp. | |||
| Postia subcaesia | |||
| Russula sp. | |||
| Skeletocutis odora | |||
| Streptobotrys sp. aff. Sympodiomyces | |||
| Trametes pubescens | RG114 | ||
| Trichophyton sp. | |||
| Trichophyton sp. | |||
| Trichophyton sp. | |||
| Trichophyton sp. | |||
| Trichophyton sp. | |||
| Tyromyces sp. |
CAN, Canada; UKR, Ukraine; ON, Ontario; EST, Estonia; CHE, Switzerland; HUN, Hungary; DEU, Germany; FRA, France; BEL, Belgium; POL, Poland.
Assay analytic sensitivity.
Genomic DNA from the P. destructans type strain, 20631-21 (Ulster Co., NY, 2008), was prepared with the OmniPrep for Fungi kit. Genomic DNA from the near relative Pseudogymnoascus sp. strain 24MN13 (10) was prepared with the DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) using the manufacturer's supplementary protocol Purification of Total DNA from Yeast Using the DNeasy Blood & Tissue Kit (15). Briefly, scrapings of fungal mycelia from a plate culture were pretreated with lyticase to weaken the cell walls before the standard DNeasy extraction protocol. We added 600 μl of sorbitol buffer (1 M sorbitol, 100 mM EDTA, 14 mM beta-mercaptoethanol) containing 200 U of lyticase to the mycelial samples and incubated them for 30 min at 30°C. Samples were centrifuged for 10 min at 300 × g, and the supernatant was discarded. The extraction was continued with the manufacturer's DNeasy extraction protocol. The lysis step was conducted for 12 h at 56°C. The P. destructans and Pseudogymnoascus sp. 24MN13 genomic DNA preparations were quantified using the Quant-IT PicoGreen double-stranded DNA (dsDNA) assay kit (Life Technologies, Carlsbad, CA) according to the manufacturer's protocol. Assay fluorescence was measured with a DynaQuant 300 fluorometer (Harvard Bioscience, Inc., Holliston, MA). DNA was diluted to approximately 10 ng/μl and quantified as described above, diluted initially to 200 pg/μl, and then serially diluted to 200 ag/μl immediately before assay. All dilutions were prepared in UltraPure water containing 0.05% Tween 20. Assays were conducted in triplicate with 4 μl of template DNA. Quantities of template DNA were plotted versus qPCR CT to produce standard curves for quantification.
Assay benchmarking.
The ITS1-qPCR was assessed relative to the α-(l)-rhamnosidase (ALR) PCR (9) and the intergenic spacer (IGS) PCR (11) for sensitivity in detecting P. destructans. The assays were run with the same conditions as the ITS1-qPCR sensitivity test described above. Specifically, the three assays were run in triplicate on the same real-time PCR instrument and plate, with the same thermocycling conditions, P. destructans DNA serial dilutions, and DNA template quantity. To minimize the variability between the three assays, the ITS1-qPCR mixture was used for all assays, with the ITS1-qPCR primers and probes replaced with those specific to the IGS or ALR assays at the concentrations recommended by their developers (9, 11). An identical test was conducted immediately after the completion of the previous one, but using the PCR mixture and thermal cycling conditions recommended for the IGS-qPCR (11) to determine if these factors have an impact upon the performance of the three assays.
Resolution of mixtures of P. destructans and other Pseudogymnoascus spp.
To determine the assay's ability to resolve mixtures of DNA from P. destructans and other Pseudogymnoascus spp. in samples, we prepared mixtures from genomic template DNA of P. destructans 20631-21 (4) and Pseudogymnoascus sp. 24MN13 (10). Genomic DNA was quantified and diluted as appropriate in the same manner as for the sensitivity analysis. Mixtures of the two genomes were prepared in 100%, 99%, 90%, 75%, 50%, 25%, 10%, 5%, 3%, 1%, and 0% proportions of P. destructans relative to Pseudogymnoascus sp. 24MN13. These mixtures were prepared at both 200 pg and 200 fg of total DNA per PCR. Mixtures were assayed in triplicate. Quantities of DNA from both species in each mixture were calculated and plotted versus the CT for the corresponding probe signal to produce standard curves for quantification.
Environmental sample assay.
A total of 5,659 environmental samples, including 5,352 bat skin swabs, 226 swabs of hibernaculum surfaces, and 81 hibernaculum sediment samples, were assayed with both the ITS1-qPCR using 2 μl of template DNA and the IGS-qPCR (11) using the master mix, thermal cycling conditions, and 5 μl of template DNA recommended by the developers. Samples were assayed at least twice with each procedure. DNA from swab samples was extracted with a DNeasy Blood & Tissue Kit using the manufacturer's supplementary yeast protocol, as described above. Sediment samples were extracted using a PowerSoil DNA isolation kit (MO BIO, Carlsbad, CA) according to the manufacturer's protocol. University of California—Santa Cruz Institutional Animal Care and Use Committee approval was obtained for sample collection from bats.
RESULTS
Assay specificity.
FAM fluorescence reached the threshold for all 35 P. destructans DNA samples in the positive-control panel, correctly identifying these samples as P. destructans. VIC fluorescence did not reach the threshold for any of these samples. For the Pseudogymnoascus and Geomyces near-relative samples in the panel, VIC fluorescence always reached the threshold, correctly differentiating them from the P. destructans clade. However, the FAM signal occasionally crossed the threshold for the near-relative samples as well. In these cases, the FAM signal always lagged behind the VIC signal, with a minimum CT difference of 11.1 between the two signals. This indicates that the P. destructans probe has some affinity for the Pseudogymnoascus/Geomyces allele. A CT difference between the VIC and FAM signals less than 11.1 likely indicates that the sample is a mixture of near-relative and P. destructans DNA. The three strains of Leuconeurospora assayed did not yield positive results in the ITS1-qPCR assay. This confirms in silico comparisons to Leuconeurospora and Pseudeurotium sequences, the closest known relatives of Pseudogymnoascus spp., which have poor similarity to the ITS1-qPCR forward primer, preventing amplification of these and other more distantly related fungi.
Assay analytic sensitivity and benchmarking.
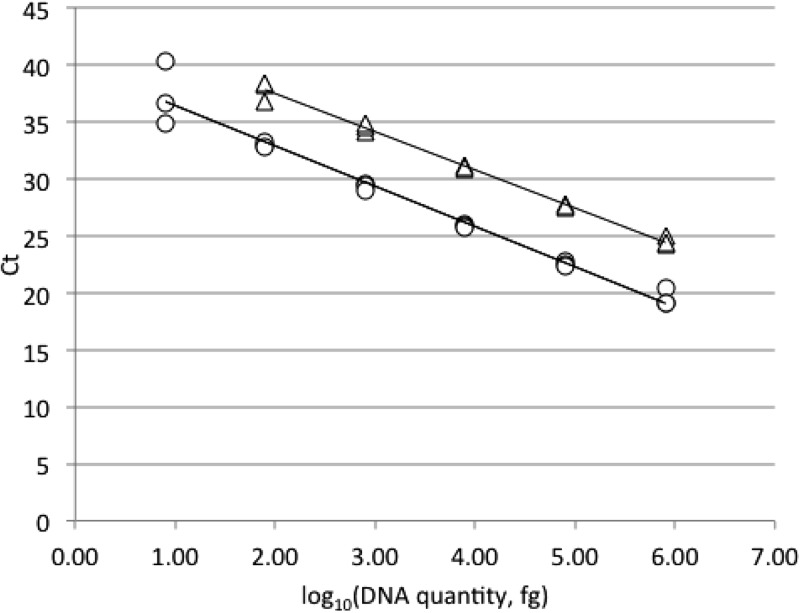
The ITS1-qPCR successfully detected 8 fg of P. destructans DNA in triplicate (average CT ± SD = 37.3 ± 2.8). The assay was quantitative for P. destructans over 5 orders of magnitude (Fig. 1). The sensitivity for Pseudogymnoascus sp. 24MN13 was an order of magnitude less—80 fg of DNA in triplicate (average CT ± SD = 37.8 ± 0.87)—and was quantitative over 4 orders of magnitude (Fig. 1). By comparison, the IGS-PCR could also detect P. destructans template DNA in triplicate at 8 fg (average CT ± SD = 38.7 ± 2.1) and once at 800 ag (CT = 40.5). The ALR-PCR detected 800 fg of P. destructans DNA in triplicate (average CT ± SD = 37.3 ± 0.35) and once at 80 fg (CT = 40.5). The CT of the IGS-PCR trailed that of the ITS1-qPCR an average of 1.18 cycles, whereas the CT of the ALR-PCR trailed that of the ITS1-qPCR an average of 8.26 cycles. This demonstrates that the sensitivity of the ITS1-qPCR is comparable to that of the IGS-PCR and over 2 orders of magnitude greater than that of the ALR-PCR. Similar results were obtained when the optimized IGS-PCR mixture and thermal cycling conditions were used (data not shown).
FIG 1.
Standard curves for the quantification of P. destructans 20631–21 (○) and Pseudogymnoascus sp. 24MN13 (△) genomic DNA with ITS1-qPCR. The relationship between CT and log10 fg of DNA for the assay is linear for P. destructans over 5 orders of magnitude (y = −3.5356x + 39.924; R2 = 0.971) and 4 orders of magnitude for Pseudogymnoascus sp. 24MN13 (y = −3.3464x + 44.134; R2 = 0.993).
Resolution of P. destructans and Pseudogymnoascus sp. 24MN13 mixtures.
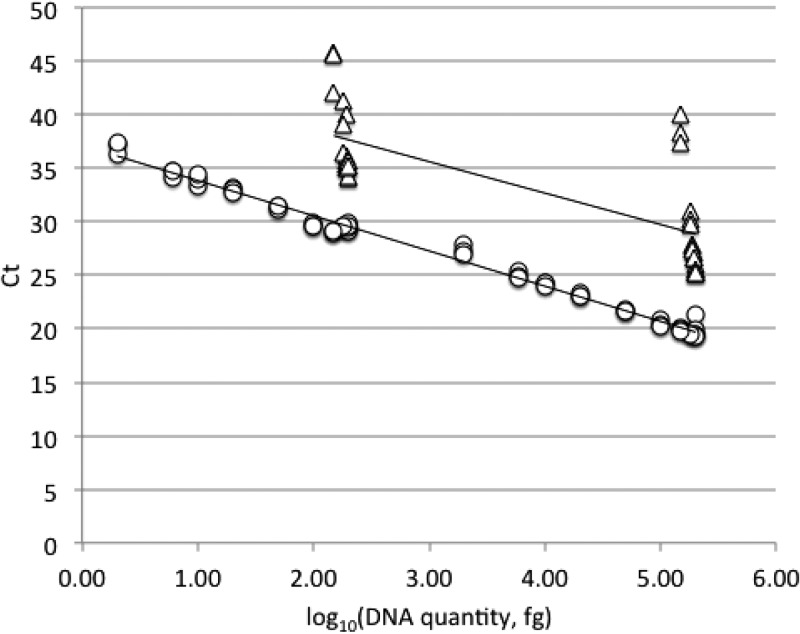
Pseudogymnoascus destructans DNA was detected in all mixtures, from proportions of 99% to 1% P. destructans. The assay was quantitative for P. destructans in all mixtures (Fig. 2). Pseudogymnoascus sp. 24MN13 could be detected only in mixtures of 99%, 97%, 95%, 90%, and 75% relative to P. destructans. ITS1-qPCR CT values were insensitive to Pseudogymnoascus sp. template quantity, and the relationship between CT and template DNA quantity varied considerably between the two total DNA concentrations tested (Fig. 2).
FIG 2.
Standard curves for the quantification of mixtures of P. destructans 20631-21 (○) and Pseudogymnoascus sp. 24MN13 (△) genomic DNA with ITS1-qPCR. The standard curve for P. destructans (y = −3.289x + 37.12; R2 = 0.990) was linear across all mixture samples. In contrast, assay CT was insensitive to Pseudogymnoascus sp. quantity (y = −2.9363x + 44.403; R2 = 0.544).
As noted in the specificity evaluation, the FAM signal occasionally crossed the threshold for pure Pseudogymnoascus sp. DNA, with a minimum observed CT difference of 11.1. Once again, in the mixtures experiment, a FAM signal was observed in the 100% Pseudogymnoascus sp. control. The average CT difference between the FAM and VIC signals was 7.86. For the mixtures, the difference between signals never exceeded this value (1.65 was the largest difference, occurring in the 200-pg 1% P. destructans sample). CT differences tended to be higher for the 200-pg template assay than for the 20-pg template assay (data not shown). It is therefore unlikely that mixtures of P. destructans and other Pseudogymnoascus spp. would be mistaken for samples containing only Pseudogymnoascus spp. unless the sample DNA concentrations were much higher than those tested in this experiment.
Environmental sample assay.
Of the 5,659 environmental samples tested, 1,200 were determined to be positive for P. destructans with the ITS1-qPCR assay, and 1,613 were found to be positive with the IGS-qPCR. Sample-by-sample comparisons of the two assays are not appropriate, however, due to the larger amount of template used in the IGS-qPCR (5 μl) than in the ITS1-qPCR (2 μl). Exact correspondence between the two assays is also unlikely because most of the positive qPCR samples had a very high CT (greater than 38), indicating a very low DNA concentration near the detection limit of both assays. Nonetheless, the two assays agreed on 3,616 of 3,617 (99.97%) of negative results.
DISCUSSION
Three real-time PCR assays for P. destructans have been developed to date: the ALR-PCR (9), the IGS-PCR (11), and the ITS1-qPCR described in this paper. Due to the similarity of P. destructans to other Pseudogymnoascus species known to inhabit the same environments (10), assay specificity is essential. The ITS1-qPCR was screened successfully against a diverse panel of North American and European P. destructans isolates from infected bats, other North American Pseudogymnoascus sp. isolates obtained from bat hibernacula and genotyped by ITS1 sequencing (10), and related Geomyces and Leuconeurospora strains, as well as a variety of other fungi (Table 1). The specificity of our assay for P. destructans, other Pseudogymnoascus spp., and Geomyces spp. outside North America and Europe is currently unknown, as is the full extent of diversity in these common but poorly understood taxa. We are actively pursuing additional worldwide isolates from the family Pseudeurotiaceae to address this knowledge gap.
Assay sensitivity is critical to detect potentially low target DNA quantities, particularly in samples from low-intensity infections or those with a low signal-to-noise ratio. We demonstrated that the ITS1-qPCR can detect 8 fg of P. destructans genomic DNA, comparable to the IGS-PCR and at least 100 times more sensitive than the ALR-PCR. This is likely due to the fact that the targets for both the ITS1-qPCR and IGS-PCR lie on the multicopy rRNA operon, whereas the ALR targets a single-copy gene. The P. destructans genome contains approximately 26 copies of the rRNA operon based on the 30.65-Mb whole-genome sequence assembly (Geomyces destructans Sequencing Project, Broad Institute, Cambridge, MA [http://www.broadinstitute.org/]). The 8-fg detection limit corresponds to less than 1 copy (∼0.26 copies) of the haploid genome.
The ITS1-qPCR is the first reported qPCR capable of detecting non-destructans Pseudogymnoascus and Geomyces species. Members of Geomyces have been previously detected in the microbial communities of cold high-altitude or high-latitude soils, terrestrial and aquatic plants, surface waters (freshwater and marine), deep-sea communities, and ice (16, 17), keratin-rich environments such as decaying fur and feathers (18), and skin infections (19). The emergence of WNS has sparked an interest in the distribution and ecology of Pseudogymnoascus and Geomyces species in relation to their pathogenic relatives (10, 14). Recently, the ectomycota of hibernating bats was sampled with culture-based methods (20, 21), revealing the presence of Geomyces and Leuconeurospora on bats and corroborating previous work on hibernaculum sediment (10). Clearly, there is much to be learned about the fungal communities of bats and their hibernation sites. Our ITS1-qPCR should serve as a useful tool in such studies of these relatively unknown but likely ubiquitous genera.
In conclusion, we present the development of a sensitive, high-throughput method for identifying and differentiating P. destructans and near-relative fungi in DNA extracts from culture and various environmental samples. The specificity of the assay coupled with its sensitivity and flexibility support its utility as an essential tool for both research and risk assessment of the devastating bat epizootic white-nose syndrome, as well as ecological studies of the genera Pseudogymnoascus and Geomyces.
ACKNOWLEDGMENTS
We thank Katy Parise, Andrew Minnis, Andrea Gargas, Nicolette Janke, Christy Allen, Sabrina German, Colin Sobek, Joe Okoniewski, Kate Langwig, and Andrew Krohn for their invaluable aid in developing this protocol. DNA samples critical to the validation of the qPCR were provided by David Blehert, Jeffrey Lorch, Gudrun Wibbelt, and Sébastien Puechmaille. The manuscript and analyses were improved by discussions with Marm Kilpatrick and Winifred Frick.
This work was supported by funding from the U.S. Fish & Wildlife Service (grant number 50120-B-G004), Bat Conservation International, and the National Science Foundation (DEB-1115895).
Footnotes
Published ahead of print 27 December 2013
REFERENCES
- 1.Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JTH, Darling SR, Gargas A, Niver R, Okoniewski JC, Rudd RJ, Stone WB. 2009. Bat white-nose syndrome: an emerging fungal pathogen? Science 323:227. 10.1126/science.1163874 [DOI] [PubMed] [Google Scholar]
- 2.US Fish and Wildlife Service 17 January 2012. North American bat death toll exceeds 5.5 million from white-nose syndrome. US Fish & Wildlife Service news release. http://www.fws.gov/northeast/feature_archive/Feature.cfm?id=794592078 [Google Scholar]
- 3.Lorch JM, Meteyer CU, Behr MJ, Boyles JG, Cryan PM, Hicks AC, Ballmann AE, Coleman JTH, Redell DN, Reeder DM, Blehert DS. 2011. Experimental infection of bats with Geomyces destructans causes white-nose syndrome. Nature 480:376–378. 10.1038/nature10590 [DOI] [PubMed] [Google Scholar]
- 4.Gargas A, Trest MT, Christensen M, Volk TJ, Blehert DS. 2009. Geomyces destructans sp. nov. associated with bat white-nose syndrome. Mycotaxon 108:147–154. 10.5248/108.147 [DOI] [Google Scholar]
- 5.Meteyer CU, Buckles EL, Blehert DS, Hicks AC, Green DE, Shearn-Bochsler V, Thomas NJ, Gargas A, Behr MJ. 2009. Histopathologic criteria to confirm white-nose syndrome in bats. J. Vet. Diagn. Invest. 21:411–414. 10.1177/104063870902100401 [DOI] [PubMed] [Google Scholar]
- 6.Lorch JM, Muller LK, Russel RE, O'Connor M, Lindner DL, Blehert DS. 2013. Distribution and environmental persistence of the causative agent of white-nose syndrome, Geomyces destructans, in bat hibernacula of the eastern United States. Appl. Environ. Microbiol. 79:1293–1301. 10.1128/AEM.02939-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturvedi V, Springer DJ, Behr MJ, Ramani R, Li XJ, Peck MK, Ren P, Bopp DJ, Wood B, Samsonoff WA, Butchkoski CM, Hicks AC, Stone WB, Rudd RJ, Chaturvedi S. 2010. Morphological and molecular characterizations of psychrophilic fungus Geomyces destructans from New York bats with white nose syndrome (WNS). PLoS One 5:e10783. 10.1371/journal.pone.0010783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorch JM, Gargas A, Meteyer CU, Berlowski-Zier BM, Green DE, Shearn-Bochsler V, Thomas NJ, Blehert DS. 2010. Rapid polymerase chain reaction diagnosis of white-nose syndrome in bats. J. Vet. Diagn. Invest. 22:224–230. 10.1177/104063871002200208 [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi S, Rudd RJ, Davis A, Victor TR, Li XJ, Appler KA, Rajkumar SS, Chaturvedi V. 2011. Rapid real-time PCR assay for culture and tissue identification of Geomyces destructans: the etiologic agent of bat geomycosis (white nose syndrome). Mycopathologia 172:247–256. 10.1007/s11046-011-9435-5 [DOI] [PubMed] [Google Scholar]
- 10.Lindner DL, Gargas A, Lorch JM, Banik MT, Glaeser J, Kunz TH, Blehert DS. 2011. DNA-based detection of the fungal pathogen Geomyces destructans in soils from bat hibernacula. Mycologia 103:241–246. 10.3852/10-262 [DOI] [PubMed] [Google Scholar]
- 11.Muller LK, Lorch JM, Lindner DL, O'Connor M, Gargas A, Blehert DS. 2013. Bat white-nose syndrome: a real-time TaqMan polymerase chain reaction test targeting the intergenic spacer region of Geomyces destructans. Mycologia 105:253–259. 10.3852/12-242 [DOI] [PubMed] [Google Scholar]
- 12.Minnis AM, Lindner DL. 2013. Phylogenetic evaluation of Geomyces and allies reveals no close relatives of Pseudogymnoascus destructans, comb. nov., in bat hibernacula of eastern North America. Fungal Biol. 117:638–649. 10.1016/j.funbio.2013.07.001 [DOI] [PubMed] [Google Scholar]
- 13.Ganley ARD, Kobayashi T. 2007. Highly efficient concerted evolution in the ribosomal DNA repeats: total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 17:184–191. 10.1101/gr.5457707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorch JM, Lindner DL, Gargas A, Muller LK, Minnis AM, Blehert DS. 2013. A culture-based survey of fungi in soil from bat hibernacula in the eastern United States and its implications for detection of Geomyces destructans, the causal agent of bat white-nose syndrome. Mycologia 105:237–252. 10.3852/12-207 [DOI] [PubMed] [Google Scholar]
- 15.Qiagen 2006. Purification of total DNA from yeast using the DNeasy Blood & Tissue Kit (DY13 Aug-06). Qiagen, Hilden, Germany [Google Scholar]
- 16.Hayes MA. 2012. The Geomyces fungi: ecology and distribution. Bioscience 62:819–823. 10.1525/bio.2012.62.9.7 [DOI] [Google Scholar]
- 17.Gonçalves VN, Vaz ABM, Rosa CA, Rosa LH. 2012. Diversity and distribution of fungal communities in lakes of Antarctica. FEMS Microbiol. Ecol. 82:459–471. 10.1111/j.1574-6941.2012.01424.x [DOI] [PubMed] [Google Scholar]
- 18.Marshall WA. 1998. Aerial transport of keratinaceous substrate and distribution of the fungus Geomyces pannorum in Antarctic soils. Microb. Ecol. 36:212–219. 10.1007/s002489900108 [DOI] [PubMed] [Google Scholar]
- 19.Gianni C, Caretta G, Romano C. 2003. Skin infection due to Geomyces pannorum var. pannorum. Mycoses 46:430–432. 10.1046/j.1439-0507.2003.00897.x [DOI] [PubMed] [Google Scholar]
- 20.Vanderwolf KJ, McAlpine DF, Malloch D, Forbes GJ. 2013. Ectomycota associated with hibernating bats in eastern Canadian caves prior to the emergence of white-nose syndrome. Northeast Nat. 20:115–130. 10.1656/045.020.0109 [DOI] [Google Scholar]
- 21.Johnson LJ, Miller AN, McCleery RA, McClanahan R, Kath JA, Lueschow S, Porras-Alfaro A. 2013. Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of white nose syndrome. Appl. Environ. Microbiol. 79:5465–5471. 10.1128/AEM.01429-13 [DOI] [PMC free article] [PubMed] [Google Scholar]