Abstract
We describe here the sequence and gene organization of a new glycopeptide resistance operon (vanO) in Rhodococcus equi from soil. The vanO operon has low homology to enterococcal van operons and harbors a vanHOX cluster transcribed in the direction opposite that of the vanS-vanR regulatory system and composed of three open reading frames with unknown function. This finding has clinical interest, since glycopeptides are used to treat R. equi infections and resistance has been reported in clinical isolates.
TEXT
Glycopeptides such as vancomycin and teicoplanin are last-resort drugs for treatment of nosocomial infections caused by methicillin-resistant Staphylococcus aureus and enterococci (1). Resistance is due to synthesis of low-affinity peptidoglycan (PG) precursors that terminate with either d-Ala–d-Lac or d-Ala–d-Ser (2). In enterococci, four d-Ala–d-Lac operons (vanA, vanB, vanD, and vanM) and five d-Ala–d-Ser operons (vanC, vanE, vanG, vanL, and vanN) have been described to date (3–7). It has been hypothesized that glycopeptide resistance operons originated from glycopeptide producers and other Gram-positive bacteria residing in soil (8).
The aim of this study was to elucidate the genetic organization of glycopeptide resistance operon in Rhodococcus equi RE-S7B isolated from garden soil in Denmark in 2004 (9). Rhodococcus equi is a Gram-positive soil saprophytic coccobacillus associated with severe bronchopneumonia in foals (10–12) and rare but fatal infections in immunocompromised patients (13, 14). Vancomycin is a last-resort drug for treatment of R. equi infections in humans (13), and resistance to vancomycin has been reported in clinical isolates from Taiwan (15), but the genetic mechanism of resistance was not investigated.
Whole-genome sequencing of RE-S7B was performed using Illumina paired-end (PE) technology (500-bp library) (BGI, Hong Kong). De novo genome assembly of the reads was performed using Geneious v7.3, and putative glycopeptide resistance genes were annotated using CLC Genomics Workbench v6.8.2, NCBI BLAST, and the NCBI conserved domain search. The Illumina 500-bp PE library generated 700 Mb of data and 7,777,778 paired reads, and de novo genome assembly produced 427 contigs of ≥1,000 bp with a mean length of 18,575 bp. RE-S7B was confirmed to be R. equi based upon 98.6% 16S rRNA gene sequence identity to reference strain R. equi DSM20307. By use of the partial ligase gene sequence obtained by Guardabassi et al. (9), a putative glycopeptide resistance operon was localized on a 28,806-bp contig harboring 30 open reading frames (ORFs).
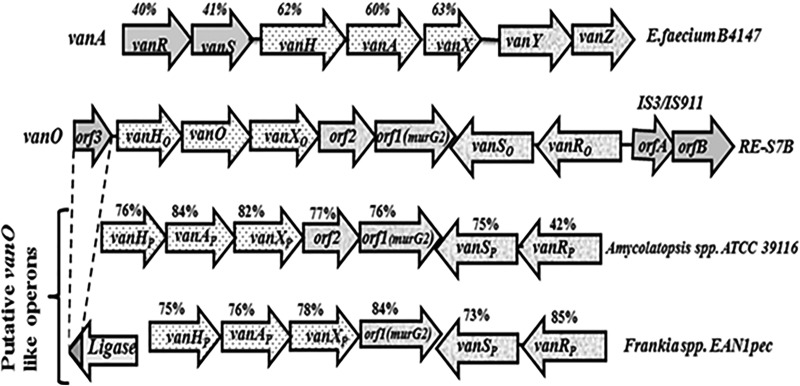
The operon in RE-S7B, designated vanO, had unique gene organization in comparison with the vanA operon in enterococci and displayed structural similarities to putative vanHAX operon-like clusters detected in actinomycete Amycolatopsis sp. strain ATCC 39116 (16) and Frankia sp. strain EAN1pec (GenBank accession no.YP_001511364) (Fig. 1). The vanO operon harbored a vanHOX resistance gene cluster transcribed in the direction opposite that of the vanS-vanR two-component regulatory system. The percentages of nucleotide sequence identity to the vanHAX gene cluster in enterococci ranged between 60 and 63%, whereas higher homology (76 to 84%) to the corresponding vanHAX clusters in the actinomycetes was observed (Fig. 1). Upstream of vanHOX, orf3 was annotated, and this was determined to be a truncated murF homologue sharing 83% predicted amino acid identity to the 3′ end of the murF-like gene described in the teicoplanin producer Actinoplanes teichomyceticus (17). Downstream of vanHOX, orf1 and orf2 were identified, and these are not similar to any genes described in the van operons known to date. Orf1 is 84% identical to a putative MurG-like protein described in Frankia sp. EAN1pec (accession no. YP_001511362), while Orf2 does not have a protein homologue in publicly available databases. Homology modeling using structure guided sequence profiles (18) showed that Orf2 is a peptidase C39-like family protein (data not shown).
FIG 1.
Organization of vanA in Enterococcus faecium (accession no. M97297), vanO in Rhodococcus equi RE-S7B (accession no. KF478993), and putative van-like clusters in Amycolatopsis sp. ATCC 39116 (accession no. WP_020417885) and Frankia sp. EAN1pec (accession no.YP_001511364). The arrows show directions of transcription. Percentages above the arrows indicate identity of the corresponding gene to vanO operon in RE-S7B. “P” refers to “putative,” to indicate van the operons in Amycolatopsis and Frankia species have never been reported, and their accession numbers refer to the putative ligases (vanAP).
We identified upstream of the vanS-vanR two-component regulatory system an IS3-like transposase gene with 83% nucleotide identity to putative IS3-like elements (orfA and orfB) in pyridine-degrading Rhodococcus sp. strain PY11 (accession numbers ZP_09309307.1 and ZP_09309306.1). IS3-like elements are known to catalyze the “figure-eight” form of transposition across bacterial species (19–21). Acquisition of glycopeptide resistance in clinically important bacteria is generally due to horizontal transfer of plasmids carrying Tn3 transposons, which contain transposases that are structurally and functionally distinct from the IS3 type (22–24). Thus, it appears that the vanO operon in RE-S7B is distantly related to and has evolved separately from van operons in enterococci. This was confirmed by the GC content (61 to 71%) of the vanO operon genes, which is higher than the GC contents of enterococcal genes in vanA operons but similar to those of genes in actinomycetes. This is not surprising, since R. equi is phylogenetically more closely related to actinomycetes than to enterococci.
Transfer of vanO-mediated glycopeptide resistance was attempted by filter mating experiments (16) using vancomycin-susceptible (Vans), rifampin-resistant (Rifr) Enterococcus faecium BM4105RF and by transformation of RE-S7B plasmid DNA into Vans Rifr R. equi SD64 (this study). All attempts to transfer glycopeptide resistance by conjugation and transformation failed. Southern hybridization was performed to investigate whether vanO was localized on the chromosome or on a plasmid. Briefly, genomic (Easy-DNA kit; Invitrogen) and plasmid (HiPure Plasmid Midiprep kit; Invitrogen) DNAs were digested with BamHI and hybridized with a digoxigenin (DIG)-labeled vanX probe according to the manufacturer's instructions (Roche Applied Science). Signals were detected from the genomic DNA only, indicating the chromosomal location of the vanO operon (see Fig. S1 in the supplemental material).
Expression of glycopeptide resistance in RE-S7B was inducible, and vancomycin MIC values increased proportionally depending on the amount of vancomycin to which cells were exposed prior to MIC determination. RE-S7B cells pregrown in brain heart infusion broth supplemented with 8 μg/ml of vancomycin grew faster in the presence of vancomycin than nonexposed cells (see Fig. S2 in the supplemental material). The vancomycin MICs measured by Etest (bioMérieux, Denmark) using colonies from agar plates without and with 8 and 20 μg/ml vancomycin were 0.5, 48, and 98 μg/ml, respectively (see Fig. S3 in the supplemental material). Altogether, these data indicate that glycopeptide resistance was induced by exposure to vancomycin. Analysis of PG precursors (25) showed that 11% percentage of peptidoglycan precursors ended with d-Ala–d-Lac and the remaining 5% of peptidoglycan precursors ended with d-Ala–d-Ala (data not shown).
In conclusion, this study describes a novel glycopeptide resistance operon with a unique structure, designated vanO. This is the 11th van operon type detected to date, the first one described in the genus Rhodococcus. The finding of a novel glycopeptide resistance determinant in R. equi has potential implications in clinical practice, since glycopeptides are used for therapy of nosocomial infections caused by this Gram-positive species and resistance has been reported in clinical isolates (13). Future work will focus on elucidating the functions of vanO-associated orf1, orf2, and orf3 genes.
Nucleotide sequence accession number.
Newly determined sequence data for the RE-S7B strain are deposited in GenBank under accession no. KF478993.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grant HEALTH-F3-2011-282004 (EvoTAR) from the European Union.
D.D.G. thanks Marco Minoia for assistance in the Southern blot experiments.
We have no transparency declarations to declare.
Footnotes
Published ahead of print 16 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01880-13.
REFERENCES
- 1.James RC, Pierce JG, Okano A, Xie J, Boger DL. 2012. Redesign of glycopeptide antibiotics: back to the future. ACS Chem. Biol. 7:797–804. 10.1021/cb300007j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh C, Fisher S, Park IS, Prahalad M, Wu Z. 1996. Bacterial resistance to vancomycin: five genes and one missing hydrogen bond tell the story. Chem. Biol. 3:21–28. 10.1016/S1074-5521(96)90079-4 [DOI] [PubMed] [Google Scholar]
- 3.Fines M, Perichon B, Reynolds P, Sahm DF, Courvalin P. 1999. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob. Agents Chemother. 43:2161–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gholizadeh Y, Courvalin P. 2000. Acquired and intrinsic glycopeptide resistance in enterococci. Int. J. Antimicrob. Agents 16:11–17. 10.1016/S0924-8579(00)00300-9 [DOI] [PubMed] [Google Scholar]
- 5.Boyd DA, Willey BM, Fawcett D, Gillani N, Mulvey MR. 2008. Molecular characterization of Enterococcus faecalis N06-0364 with low-level vancomycin resistance harboring a novel D-Ala-D-Ser gene cluster, vanL. Antimicrob. Agents Chemother. 52:2667–2672. 10.1128/AAC.01516-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu X, Lin D, Yan G, Ye X, Wu S, Guo Y, Zhu D, Hu F, Zhang Y, Wang F. 2010. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 54:4643–4647. 10.1128/AAC.01710-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebreton F, Depardieu F, Bourdon N, Fines-Guyon M, Berger P, Camiade S, Leclercq R, Courvalin P, Cattoir V. 2011. D-Ala-D-Ser VanN-type transferable vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 55:4606–4612. 10.1128/AAC.00714-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guardabassi L, Perichon B, van Heijenoort J, Blanot D, Courvalin P. 2005. Glycopeptide resistance vanA operons in Paenibacillus strains isolated from soil. Antimicrob. Agents Chemother. 49:4227–4233. 10.1128/AAC.49.10.4227-4233.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guardabassi L, Christensen H, Hasman H, Dalsgaard A. 2004. Members of the genera Paenibacillus and Rhodococcus harbor genes homologous to enterococcal glycopeptide resistance genes vanA and vanB. Antimicrob. Agents Chemother. 48:4915–4918. 10.1128/AAC.48.12.4915-4918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vengust M, Stæmpfli H, Prescott JF. 2002. Rhodococcus equi pleuropneumonia in an adult horse. Can. Vet. J. 43:706–708 [PMC free article] [PubMed] [Google Scholar]
- 11.Venner M, Rödiger A, Laemmer M, Giguère S. 2012. Failure of antimicrobial therapy to accelerate spontaneous healing of subclinical pulmonary abscesses on a farm with endemic infections caused by Rhodococcus equi. Vet. J. 192:293–298. 10.1016/j.tvjl.2011.07.004 [DOI] [PubMed] [Google Scholar]
- 12.Muscatello G. 2012. Rhodococcus equi pneumonia in the foal–part 2: diagnostics, treatment and disease management. Vet. J. 192:27–33. 10.1016/j.tvjl.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 13.Weinstock DM, Brown AE. 2002. Rhodococcus equi: an emerging pathogen. Clin. Infect. Dis. 34:1379–1385. 10.1086/340259 [DOI] [PubMed] [Google Scholar]
- 14.Jones M, Neale T, Say P, Horne J. 1989. Rhodococcus equi: an emerging opportunistic pathogen? Aust. N. Z. J. Med. 19:103–107. 10.1111/j.1445-5994.1989.tb00213.x [DOI] [PubMed] [Google Scholar]
- 15.Hsueh P, Hung C, Teng L, Yu M, Chen Y, Wang H, Luh K. 1998. Report of invasive Rhodococcus equi infections in Taiwan, with an emphasis on the emergence of multidrug-resistant strains. Clin. Infect. Dis. 27:370–375. 10.1086/514667 [DOI] [PubMed] [Google Scholar]
- 16.Davis JR, Goodwin LA, Woyke T, Teshima H, Bruce D, Detter C, Tapia R, Han S, Han J, Pitluck S. 2012. Genome sequence of Amycolatopsis sp. strain ATCC 39116, a plant biomass-degrading actinomycete. J. Bacteriol. 194:2396–2397. 10.1128/JB.00186-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serina S, Radice F, Maffioli S, Donadio S, Sosio M. 2004. Glycopeptide resistance determinants from the teicoplanin producer Actinoplanes teichomyceticus. FEMS Microbiol. Lett. 240:69–74. 10.1016/j.femsle.2004.09.017 [DOI] [PubMed] [Google Scholar]
- 18.Nielsen M, Lundegaard C, Lund O, Petersen TN. 2010. CPHmodels-3.0—remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 38:W576–W581. 10.1093/nar/gkq535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekine Y, Eisaki N, Ohtsubo E. 1994. Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol. 235:1406–1420. 10.1006/jmbi.1994.1097 [DOI] [PubMed] [Google Scholar]
- 20.Sekine Y, Aihara K, Ohtsubo E. 1999. Linearization and transposition of circular molecules of insertion sequence IS 3. J. Mol. Biol. 294:21–34. 10.1006/jmbi.1999.3181 [DOI] [PubMed] [Google Scholar]
- 21.Chandler M, Fayet O. 1993. Translational frameshifting in the control of transposition in bacteria. Mol. Microbiol. 7:497–503. 10.1111/j.1365-2958.1993.tb01140.x [DOI] [PubMed] [Google Scholar]
- 22.Leclercq R, Derlot E, Duval J, Courvalin P. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157–161. 10.1056/NEJM198807213190307 [DOI] [PubMed] [Google Scholar]
- 23.Arthur M, Molinas C, Depardieu F, Courvalin P. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quintiliani R, Courvalin P. 1996. Characterization of Tn 1547, a composite transposon flanked by the IS 16 and IS 256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1–8. 10.1016/0378-1119(96)00110-2 [DOI] [PubMed] [Google Scholar]
- 25.Reynolds PE, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. 1994. Glycopeptide resistance mediated by enterococcal transposon Tn 1546 requires production of VanX for hydrolysis of D-alanyl-D-alanine. Mol. Microbiol. 13:1065–1070. 10.1111/j.1365-2958.1994.tb00497.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.