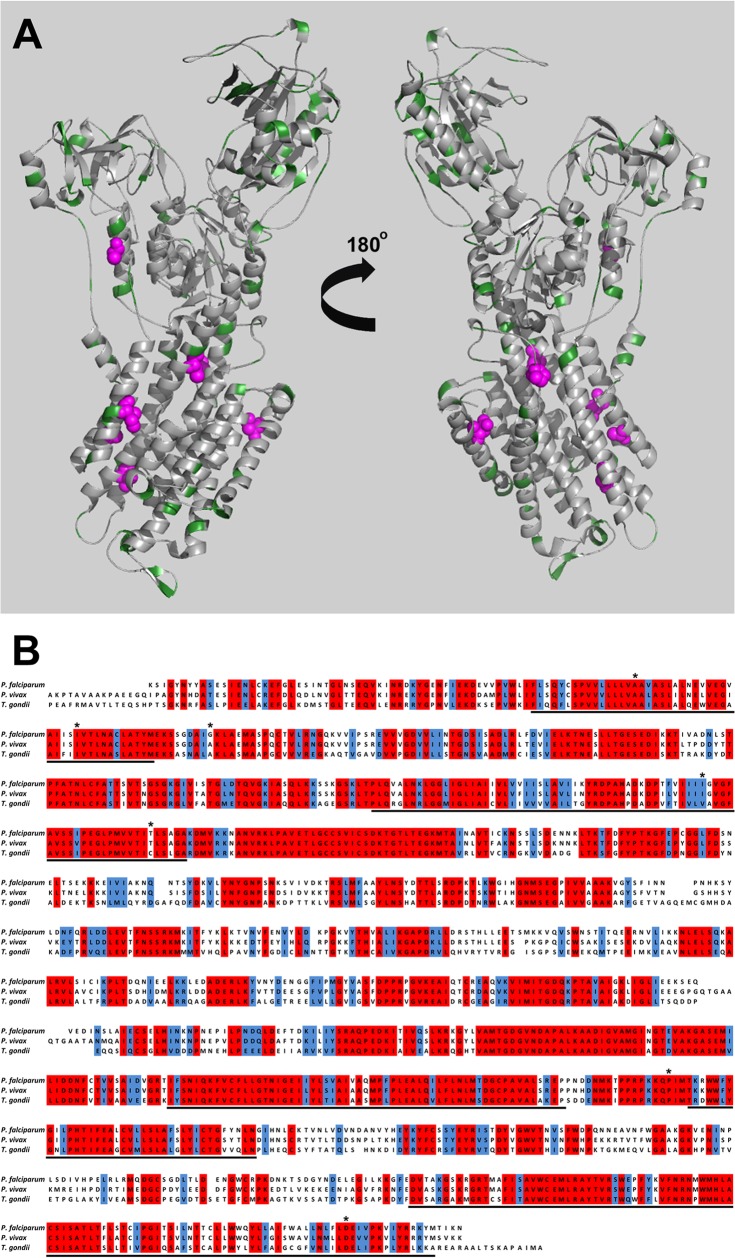
FIG 2.
(A) Modeling T. gondii ATPase4. The modeled structure of TgATPase4 is shown from the front and back in cartoon format. Residues which are significantly different in T. gondii, P. falciparum, and P. vivax are shown in green, and residues which have been shown by mutational analysis to confer spiroindolone resistance are shown as magenta spheres. The figure was produced using Pymol. (B) Multiple sequence alignment of P. falciparum, P. vivax, and T. gondii ATPase4. Residues which are fully conserved and similar are highlighted in red and blue, respectively; residues which are proposed to be associated with the membrane are underlined; and residues which confer resistance to the spiroindolone derivatives NITD678-R and NITD609-R are highlighted by asterisks.