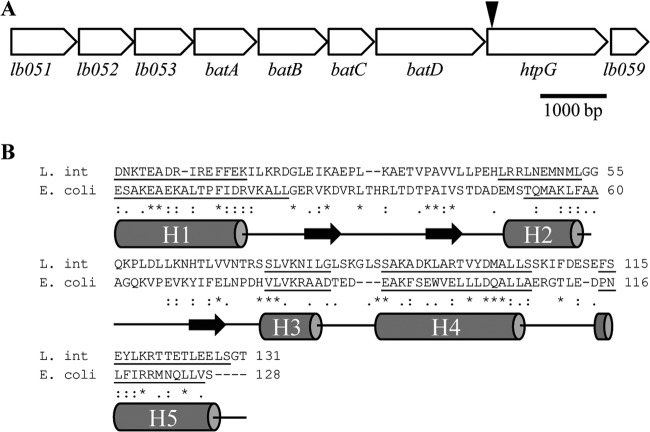
FIG 1.
Analysis of the htpG operon and protein homology. (A) Genomic organization of the putative htpG operon in L. interrogans. The gap between the coding regions of lb051 and lb052 is 6 bp. The gap between htpG and lb059 is 21 bp. The remaining genes are predicted to have overlapping open reading frames. The transposon insertion in htpG mutant M1233, located 46 bp from the start codon of htpG, is indicated by a black arrowhead. (B) C-terminal alignment of HtpG from L. interrogans (L. int) and HtpG from E. coli. The predicted secondary structure of L. interrogans HtpG shows close similarity to that of E. coli HtpG (16). Underlining indicates helical regions (H1 to H5), and black horizontal arrows represent strands, as predicted by PSIPRED (49).