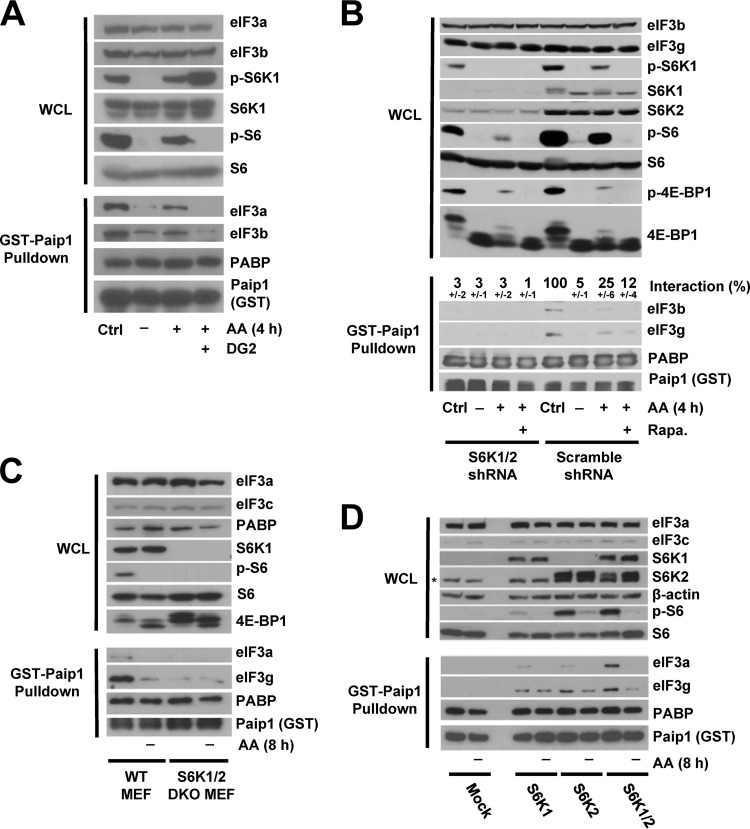
FIG 3.
Paip1-eIF3 interaction requires S6K activity. (A) HeLa cells were grown in DMEM with 10% serum (Ctrl, control condition) or amino acid starved overnight. Amino acid-starved HeLa cells were pretreated with DG2 at 5 μM (1 h) or left untreated, followed by amino acid stimulation (4 h). Whole-cell lysates (WCL) were subjected to GST-Paip1 pulldown. WCL (top panel) and GST-Paip1 pulldown eluates (bottom panel) were processed for Western blotting with the indicated antibodies: phospho-Ser240/444 S6 (p-S6) or phospho-Thr389 S6K (p-S6K). (B) HeLa cells expressing either S6K1 and S6K2 shRNAs (S6K1/2 shRNA) or Scramble shRNA were treated as in panel A using 20 nM rapamycin (Rapa.). WCL (top panel) and GST-Paip1 pulldown eluates (bottom panel) were processed for Western blotting using phospho-Ser65 4E-BP1 (p-4E-BP1). Paip1-eIF3 interaction is expressed as a percentage (±the standard deviation) of Paip1-eIF3 interaction in HeLa cells in a control condition. Quantifications were performed using ImageJ software on replicate experiments. (C) WT MEFs or S6K1/2 double-knockout MEFs were amino acid starved for 8 h and left untreated. WCL (top panel) and GST-Paip1 pulldown eluates (bottom panel) were processed for Western blotting. (D) S6K1/2 double-knockout MEFs were transduced with vector expressing S6K1, S6K2, S6K1, and S6K2 or empty vector (Mock). Cells were treated as in panel C, and WCL (top panel) and GST-Paip1 pulldown eluates (bottom panel) were processed for Western blotting. The asterisk indicates a nonspecific band.